Qingqi Hong
Scale-aware Adaptive Supervised Network with Limited Medical Annotations
Jan 02, 2026Abstract:Medical image segmentation faces critical challenges in semi-supervised learning scenarios due to severe annotation scarcity requiring expert radiological knowledge, significant inter-annotator variability across different viewpoints and expertise levels, and inadequate multi-scale feature integration for precise boundary delineation in complex anatomical structures. Existing semi-supervised methods demonstrate substantial performance degradation compared to fully supervised approaches, particularly in small target segmentation and boundary refinement tasks. To address these fundamental challenges, we propose SASNet (Scale-aware Adaptive Supervised Network), a dual-branch architecture that leverages both low-level and high-level feature representations through novel scale-aware adaptive reweight mechanisms. Our approach introduces three key methodological innovations, including the Scale-aware Adaptive Reweight strategy that dynamically weights pixel-wise predictions using temporal confidence accumulation, the View Variance Enhancement mechanism employing 3D Fourier domain transformations to simulate annotation variability, and segmentation-regression consistency learning through signed distance map algorithms for enhanced boundary precision. These innovations collectively address the core limitations of existing semi-supervised approaches by integrating spatial, temporal, and geometric consistency principles within a unified optimization framework. Comprehensive evaluation across LA, Pancreas-CT, and BraTS datasets demonstrates that SASNet achieves superior performance with limited labeled data, surpassing state-of-the-art semi-supervised methods while approaching fully supervised performance levels. The source code for SASNet is available at https://github.com/HUANGLIZI/SASNet.
A topology-preserving three-stage framework for fully-connected coronary artery extraction
Apr 02, 2025Abstract:Coronary artery extraction is a crucial prerequisite for computer-aided diagnosis of coronary artery disease. Accurately extracting the complete coronary tree remains challenging due to several factors, including presence of thin distal vessels, tortuous topological structures, and insufficient contrast. These issues often result in over-segmentation and under-segmentation in current segmentation methods. To address these challenges, we propose a topology-preserving three-stage framework for fully-connected coronary artery extraction. This framework includes vessel segmentation, centerline reconnection, and missing vessel reconstruction. First, we introduce a new centerline enhanced loss in the segmentation process. Second, for the broken vessel segments, we further propose a regularized walk algorithm to integrate distance, probabilities predicted by a centerline classifier, and directional cosine similarity, for reconnecting the centerlines. Third, we apply implicit neural representation and implicit modeling, to reconstruct the geometric model of the missing vessels. Experimental results show that our proposed framework outperforms existing methods, achieving Dice scores of 88.53\% and 85.07\%, with Hausdorff Distances (HD) of 1.07mm and 1.63mm on ASOCA and PDSCA datasets, respectively. Code will be available at https://github.com/YH-Qiu/CorSegRec.
STPNet: Scale-aware Text Prompt Network for Medical Image Segmentation
Apr 02, 2025Abstract:Accurate segmentation of lesions plays a critical role in medical image analysis and diagnosis. Traditional segmentation approaches that rely solely on visual features often struggle with the inherent uncertainty in lesion distribution and size. To address these issues, we propose STPNet, a Scale-aware Text Prompt Network that leverages vision-language modeling to enhance medical image segmentation. Our approach utilizes multi-scale textual descriptions to guide lesion localization and employs retrieval-segmentation joint learning to bridge the semantic gap between visual and linguistic modalities. Crucially, STPNet retrieves relevant textual information from a specialized medical text repository during training, eliminating the need for text input during inference while retaining the benefits of cross-modal learning. We evaluate STPNet on three datasets: COVID-Xray, COVID-CT, and Kvasir-SEG. Experimental results show that our vision-language approach outperforms state-of-the-art segmentation methods, demonstrating the effectiveness of incorporating textual semantic knowledge into medical image analysis. The code has been made publicly on https://github.com/HUANGLIZI/STPNet.
An Intra- and Cross-frame Topological Consistency Scheme for Semi-supervised Atherosclerotic Coronary Plaque Segmentation
Jan 14, 2025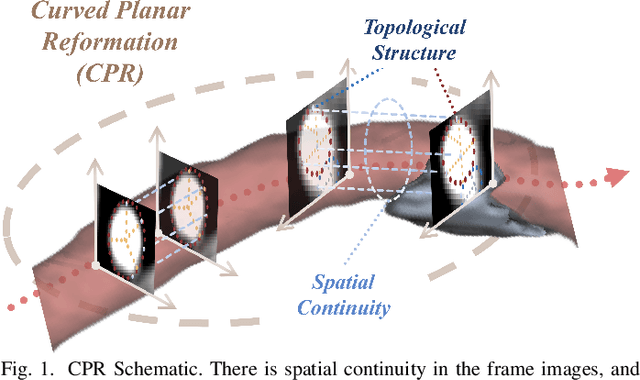
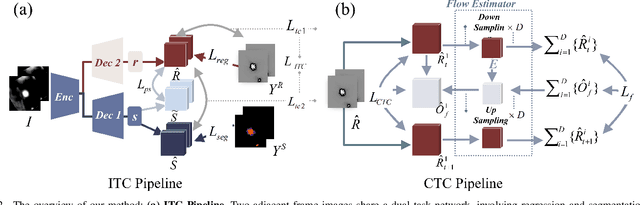
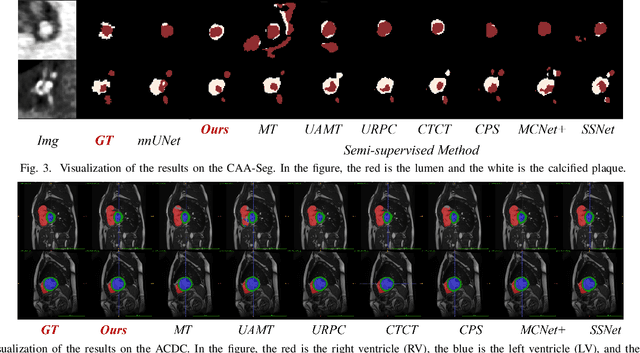
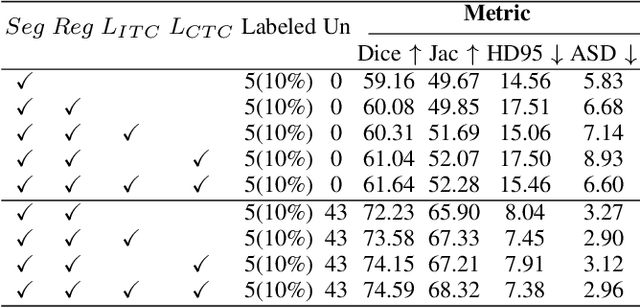
Abstract:Enhancing the precision of segmenting coronary atherosclerotic plaques from CT Angiography (CTA) images is pivotal for advanced Coronary Atherosclerosis Analysis (CAA), which distinctively relies on the analysis of vessel cross-section images reconstructed via Curved Planar Reformation. This task presents significant challenges due to the indistinct boundaries and structures of plaques and blood vessels, leading to the inadequate performance of current deep learning models, compounded by the inherent difficulty in annotating such complex data. To address these issues, we propose a novel dual-consistency semi-supervised framework that integrates Intra-frame Topological Consistency (ITC) and Cross-frame Topological Consistency (CTC) to leverage labeled and unlabeled data. ITC employs a dual-task network for simultaneous segmentation mask and Skeleton-aware Distance Transform (SDT) prediction, achieving similar prediction of topology structure through consistency constraint without additional annotations. Meanwhile, CTC utilizes an unsupervised estimator for analyzing pixel flow between skeletons and boundaries of adjacent frames, ensuring spatial continuity. Experiments on two CTA datasets show that our method surpasses existing semi-supervised methods and approaches the performance of supervised methods on CAA. In addition, our method also performs better than other methods on the ACDC dataset, demonstrating its generalization.
HAUR: Human Annotation Understanding and Recognition Through Text-Heavy Images
Dec 24, 2024Abstract:Vision Question Answering (VQA) tasks use images to convey critical information to answer text-based questions, which is one of the most common forms of question answering in real-world scenarios. Numerous vision-text models exist today and have performed well on certain VQA tasks. However, these models exhibit significant limitations in understanding human annotations on text-heavy images. To address this, we propose the Human Annotation Understanding and Recognition (HAUR) task. As part of this effort, we introduce the Human Annotation Understanding and Recognition-5 (HAUR-5) dataset, which encompasses five common types of human annotations. Additionally, we developed and trained our model, OCR-Mix. Through comprehensive cross-model comparisons, our results demonstrate that OCR-Mix outperforms other models in this task. Our dataset and model will be released soon .
Self-Supervised Learning of Deviation in Latent Representation for Co-speech Gesture Video Generation
Sep 26, 2024



Abstract:Gestures are pivotal in enhancing co-speech communication. While recent works have mostly focused on point-level motion transformation or fully supervised motion representations through data-driven approaches, we explore the representation of gestures in co-speech, with a focus on self-supervised representation and pixel-level motion deviation, utilizing a diffusion model which incorporates latent motion features. Our approach leverages self-supervised deviation in latent representation to facilitate hand gestures generation, which are crucial for generating realistic gesture videos. Results of our first experiment demonstrate that our method enhances the quality of generated videos, with an improvement from 2.7 to 4.5% for FGD, DIV, and FVD, and 8.1% for PSNR, 2.5% for SSIM over the current state-of-the-art methods.
ScribFormer: Transformer Makes CNN Work Better for Scribble-based Medical Image Segmentation
Feb 03, 2024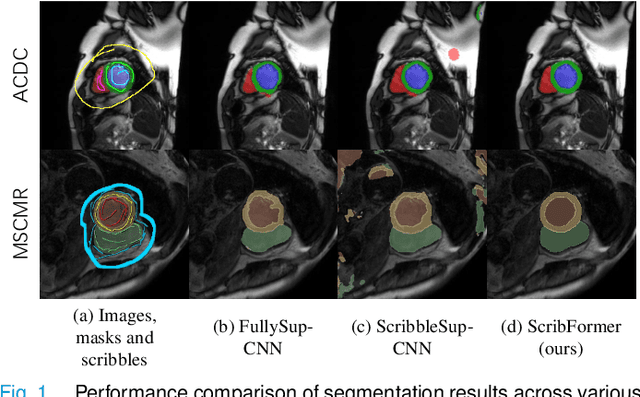
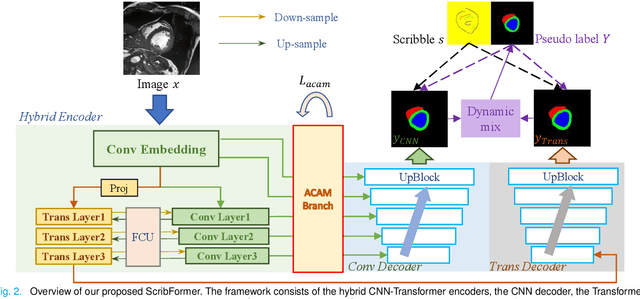
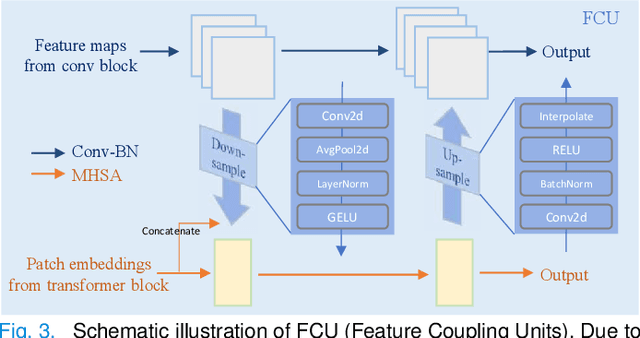
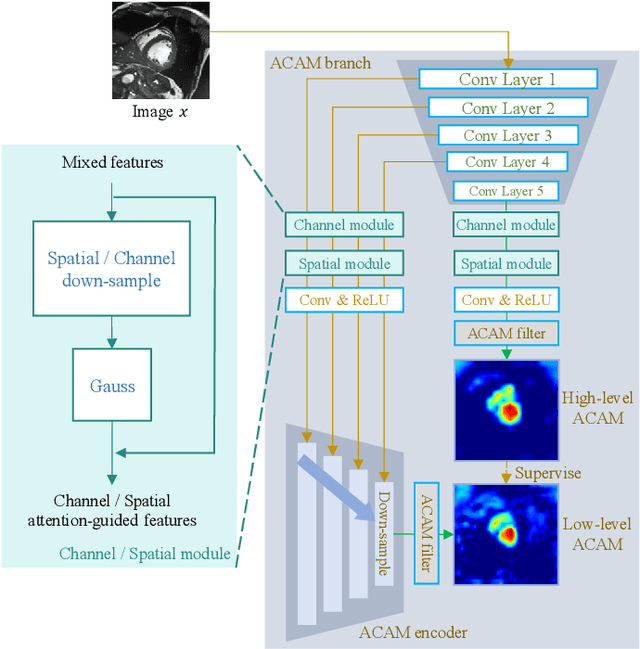
Abstract:Most recent scribble-supervised segmentation methods commonly adopt a CNN framework with an encoder-decoder architecture. Despite its multiple benefits, this framework generally can only capture small-range feature dependency for the convolutional layer with the local receptive field, which makes it difficult to learn global shape information from the limited information provided by scribble annotations. To address this issue, this paper proposes a new CNN-Transformer hybrid solution for scribble-supervised medical image segmentation called ScribFormer. The proposed ScribFormer model has a triple-branch structure, i.e., the hybrid of a CNN branch, a Transformer branch, and an attention-guided class activation map (ACAM) branch. Specifically, the CNN branch collaborates with the Transformer branch to fuse the local features learned from CNN with the global representations obtained from Transformer, which can effectively overcome limitations of existing scribble-supervised segmentation methods. Furthermore, the ACAM branch assists in unifying the shallow convolution features and the deep convolution features to improve model's performance further. Extensive experiments on two public datasets and one private dataset show that our ScribFormer has superior performance over the state-of-the-art scribble-supervised segmentation methods, and achieves even better results than the fully-supervised segmentation methods. The code is released at https://github.com/HUANGLIZI/ScribFormer.
ScribbleVC: Scribble-supervised Medical Image Segmentation with Vision-Class Embedding
Jul 30, 2023Abstract:Medical image segmentation plays a critical role in clinical decision-making, treatment planning, and disease monitoring. However, accurate segmentation of medical images is challenging due to several factors, such as the lack of high-quality annotation, imaging noise, and anatomical differences across patients. In addition, there is still a considerable gap in performance between the existing label-efficient methods and fully-supervised methods. To address the above challenges, we propose ScribbleVC, a novel framework for scribble-supervised medical image segmentation that leverages vision and class embeddings via the multimodal information enhancement mechanism. In addition, ScribbleVC uniformly utilizes the CNN features and Transformer features to achieve better visual feature extraction. The proposed method combines a scribble-based approach with a segmentation network and a class-embedding module to produce accurate segmentation masks. We evaluate ScribbleVC on three benchmark datasets and compare it with state-of-the-art methods. The experimental results demonstrate that our method outperforms existing approaches in terms of accuracy, robustness, and efficiency. The datasets and code are released on GitHub.
Coarse-to-Fine Covid-19 Segmentation via Vision-Language Alignment
Mar 01, 2023Abstract:Segmentation of COVID-19 lesions can assist physicians in better diagnosis and treatment of COVID-19. However, there are few relevant studies due to the lack of detailed information and high-quality annotation in the COVID-19 dataset. To solve the above problem, we propose C2FVL, a Coarse-to-Fine segmentation framework via Vision-Language alignment to merge text information containing the number of lesions and specific locations of image information. The introduction of text information allows the network to achieve better prediction results on challenging datasets. We conduct extensive experiments on two COVID-19 datasets including chest X-ray and CT, and the results demonstrate that our proposed method outperforms other state-of-the-art segmentation methods.
TFCNs: A CNN-Transformer Hybrid Network for Medical Image Segmentation
Jul 07, 2022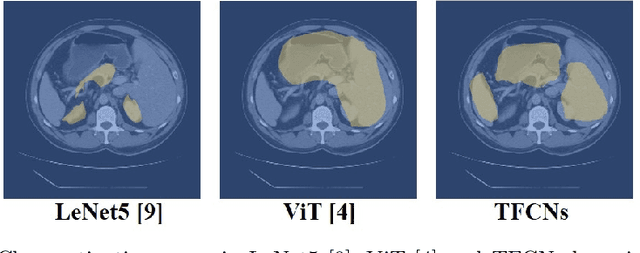
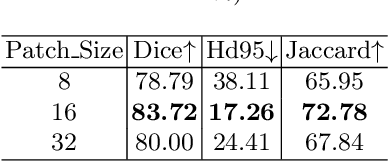
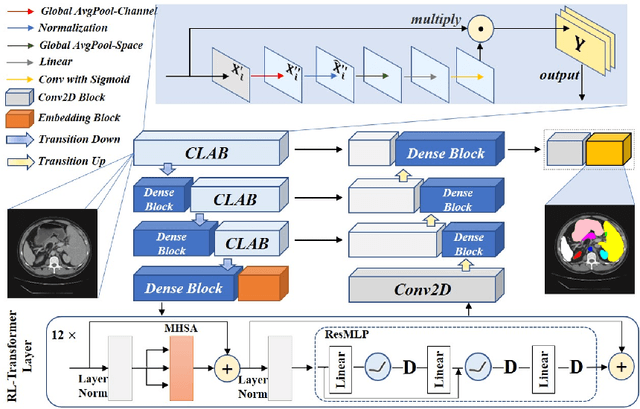
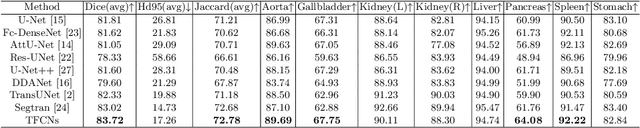
Abstract:Medical image segmentation is one of the most fundamental tasks concerning medical information analysis. Various solutions have been proposed so far, including many deep learning-based techniques, such as U-Net, FC-DenseNet, etc. However, high-precision medical image segmentation remains a highly challenging task due to the existence of inherent magnification and distortion in medical images as well as the presence of lesions with similar density to normal tissues. In this paper, we propose TFCNs (Transformers for Fully Convolutional denseNets) to tackle the problem by introducing ResLinear-Transformer (RL-Transformer) and Convolutional Linear Attention Block (CLAB) to FC-DenseNet. TFCNs is not only able to utilize more latent information from the CT images for feature extraction, but also can capture and disseminate semantic features and filter non-semantic features more effectively through the CLAB module. Our experimental results show that TFCNs can achieve state-of-the-art performance with dice scores of 83.72\% on the Synapse dataset. In addition, we evaluate the robustness of TFCNs for lesion area effects on the COVID-19 public datasets. The Python code will be made publicly available on https://github.com/HUANGLIZI/TFCNs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge