Yuchen Xu
CenBRAIN Neurotech Center of Excellence, School of Engineering, Westlake University, China, Key Laboratory of Biomedical Engineering of Ministry of Education, Qiushi Academy for Advanced Studies, Zhejiang University, China
Graph Representations for Reading Comprehension Analysis using Large Language Model and Eye-Tracking Biomarker
Jul 16, 2025Abstract:Reading comprehension is a fundamental skill in human cognitive development. With the advancement of Large Language Models (LLMs), there is a growing need to compare how humans and LLMs understand language across different contexts and apply this understanding to functional tasks such as inference, emotion interpretation, and information retrieval. Our previous work used LLMs and human biomarkers to study the reading comprehension process. The results showed that the biomarkers corresponding to words with high and low relevance to the inference target, as labeled by the LLMs, exhibited distinct patterns, particularly when validated using eye-tracking data. However, focusing solely on individual words limited the depth of understanding, which made the conclusions somewhat simplistic despite their potential significance. This study used an LLM-based AI agent to group words from a reading passage into nodes and edges, forming a graph-based text representation based on semantic meaning and question-oriented prompts. We then compare the distribution of eye fixations on important nodes and edges. Our findings indicate that LLMs exhibit high consistency in language understanding at the level of graph topological structure. These results build on our previous findings and offer insights into effective human-AI co-learning strategies.
TrafficKAN-GCN: Graph Convolutional-based Kolmogorov-Arnold Network for Traffic Flow Optimization
Mar 05, 2025Abstract:Urban traffic optimization is critical for improving transportation efficiency and alleviating congestion, particularly in large-scale dynamic networks. Traditional methods, such as Dijkstra's and Floyd's algorithms, provide effective solutions in static settings, but they struggle with the spatial-temporal complexity of real-world traffic flows. In this work, we propose TrafficKAN-GCN, a hybrid deep learning framework combining Kolmogorov-Arnold Networks (KAN) with Graph Convolutional Networks (GCN), designed to enhance urban traffic flow optimization. By integrating KAN's adaptive nonlinear function approximation with GCN's spatial graph learning capabilities, TrafficKAN-GCN captures both complex traffic patterns and topological dependencies. We evaluate the proposed framework using real-world traffic data from the Baltimore Metropolitan area. Compared with baseline models such as MLP-GCN, standard GCN, and Transformer-based approaches, TrafficKAN-GCN achieves competitive prediction accuracy while demonstrating improved robustness in handling noisy and irregular traffic data. Our experiments further highlight the framework's ability to redistribute traffic flow, mitigate congestion, and adapt to disruptive events, such as the Francis Scott Key Bridge collapse. This study contributes to the growing body of work on hybrid graph learning for intelligent transportation systems, highlighting the potential of combining KAN and GCN for real-time traffic optimization. Future work will focus on reducing computational overhead and integrating Transformer-based temporal modeling for enhanced long-term traffic prediction. The proposed TrafficKAN-GCN framework offers a promising direction for data-driven urban mobility management, balancing predictive accuracy, robustness, and computational efficiency.
Poststroke rehabilitative mechanisms in individualized fatigue level-controlled treadmill training -- a Rat Model Study
Feb 20, 2025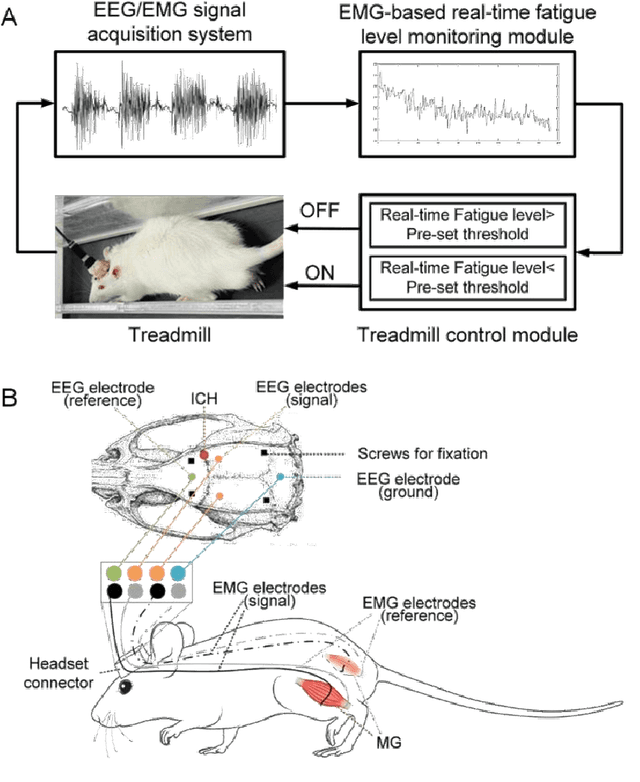
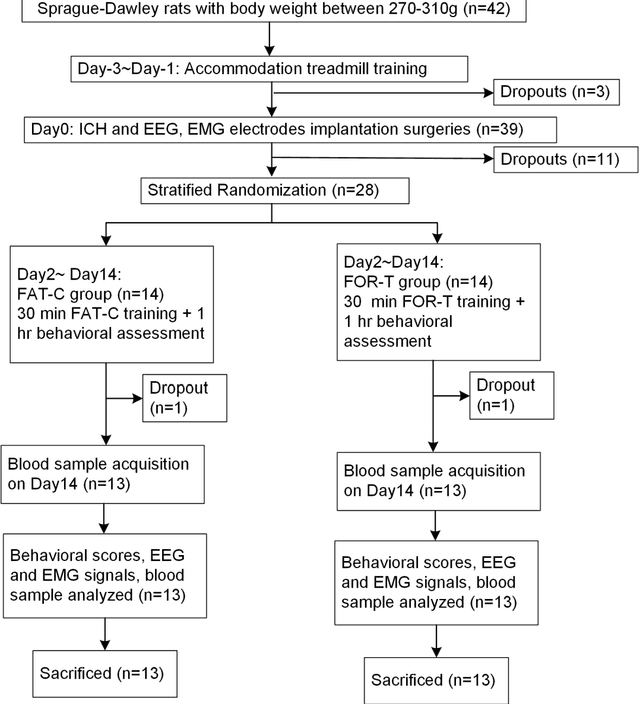
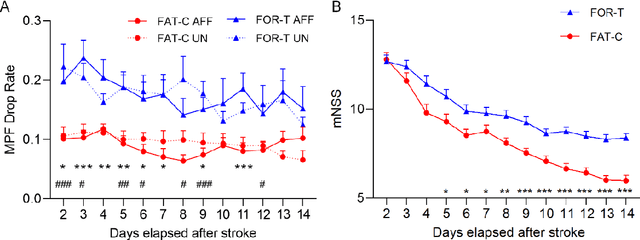
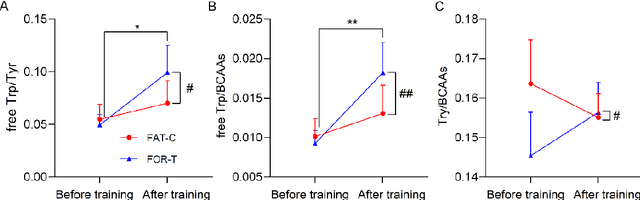
Abstract:Individualized training improved post-stroke motor function rehabilitation efficiency. However, the mechanisms of how individualized training facilitates recovery is not clear. This study explored the cortical and corticomuscular rehabilitative effects in post-stroke motor function recovery during individualized training. Sprague-Dawley rats with intracerebral hemorrhage (ICH) were randomly distributed into two groups: forced training (FOR-T, n=13) and individualized fatigue-controlled training (FAT-C, n=13) to receive training respectively from day 2 to day 14 post-stroke. The FAT-C group exhibited superior motor function recovery and less central fatigue compared to the FOR-T group. EEG PSD slope analysis demonstrated a better inter-hemispheric balance in FAT-C group compare to the FOR-T group. The dCMC analysis indicated that training-induced fatigue led to a short-term down-regulation of descending corticomuscular coherence (dCMC) and an up-regulation of ascending dCMC. In the long term, excessive fatigue hindered the recovery of descending control in the affected hemisphere. The individualized strategy of peripheral fatigue-controlled training achieved better motor function recovery, which could be attributed to the mitigation of central fatigue, optimization of inter-hemispheric balance and enhancement of descending control in the affected hemisphere.
Self-supervised Anomaly Detection Pretraining Enhances Long-tail ECG Diagnosis
Aug 30, 2024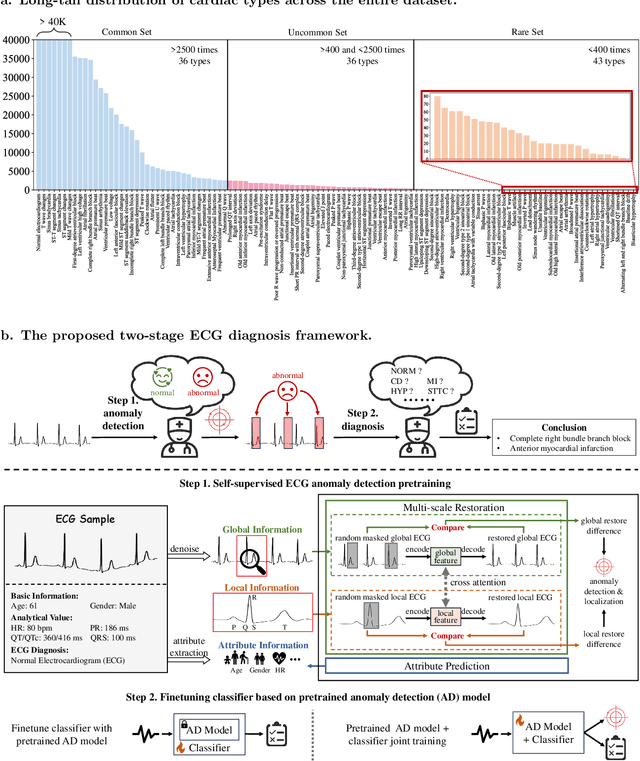
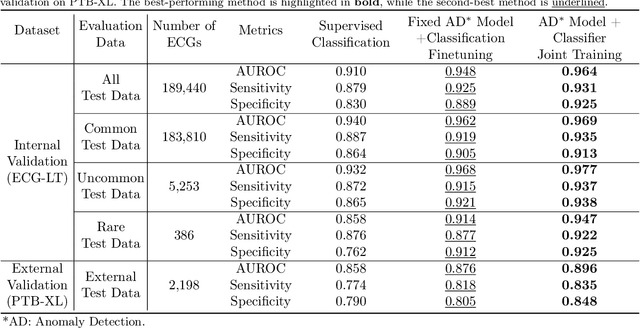
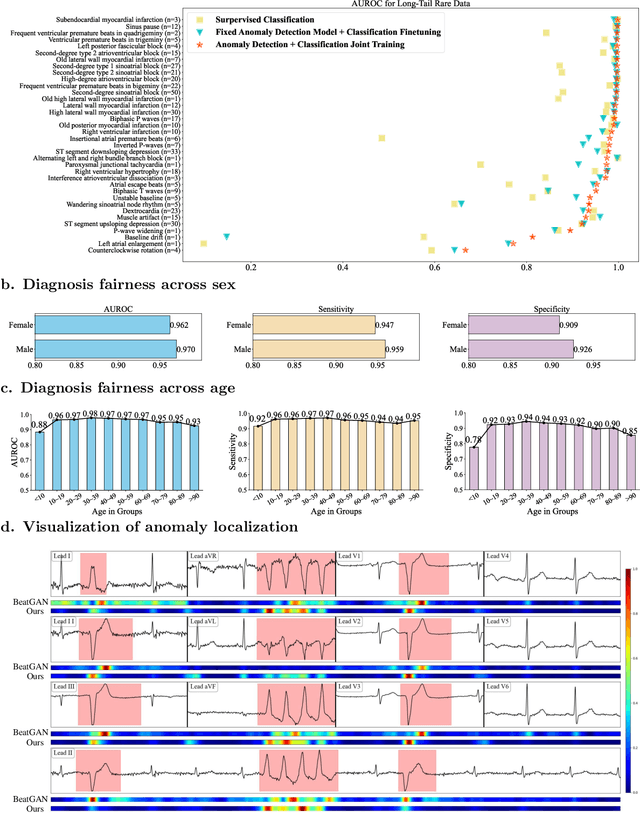
Abstract:Current computer-aided ECG diagnostic systems struggle with the underdetection of rare but critical cardiac anomalies due to the imbalanced nature of ECG datasets. This study introduces a novel approach using self-supervised anomaly detection pretraining to address this limitation. The anomaly detection model is specifically designed to detect and localize subtle deviations from normal cardiac patterns, capturing the nuanced details essential for accurate ECG interpretation. Validated on an extensive dataset of over one million ECG records from clinical practice, characterized by a long-tail distribution across 116 distinct categories, the anomaly detection-pretrained ECG diagnostic model has demonstrated a significant improvement in overall accuracy. Notably, our approach yielded a 94.7% AUROC, 92.2% sensitivity, and 92.5\% specificity for rare ECG types, significantly outperforming traditional methods and narrowing the performance gap with common ECG types. The integration of anomaly detection pretraining into ECG analysis represents a substantial contribution to the field, addressing the long-standing challenge of long-tail data distributions in clinical diagnostics. Furthermore, prospective validation in real-world clinical settings revealed that our AI-driven approach enhances diagnostic efficiency, precision, and completeness by 32%, 6.7%, and 11.8% respectively, when compared to standard practices. This advancement marks a pivotal step forward in the integration of AI within clinical cardiology, with particularly profound implications for emergency care, where rapid and accurate ECG interpretation is crucial. The contributions of this study not only push the boundaries of current ECG diagnostic capabilities but also lay the groundwork for more reliable and accessible cardiovascular care.
Anomaly Detection in Electrocardiograms: Advancing Clinical Diagnosis Through Self-Supervised Learning
Apr 07, 2024



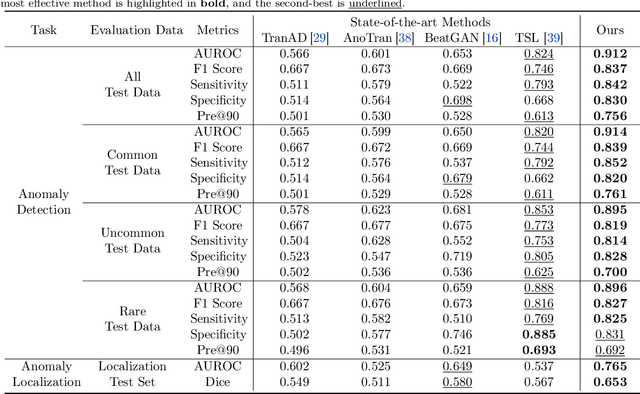
Abstract:The electrocardiogram (ECG) is an essential tool for diagnosing heart disease, with computer-aided systems improving diagnostic accuracy and reducing healthcare costs. Despite advancements, existing systems often miss rare cardiac anomalies that could be precursors to serious, life-threatening issues or alterations in the cardiac macro/microstructure. We address this gap by focusing on self-supervised anomaly detection (AD), training exclusively on normal ECGs to recognize deviations indicating anomalies. We introduce a novel self-supervised learning framework for ECG AD, utilizing a vast dataset of normal ECGs to autonomously detect and localize cardiac anomalies. It proposes a novel masking and restoration technique alongside a multi-scale cross-attention module, enhancing the model's ability to integrate global and local signal features. The framework emphasizes accurate localization of anomalies within ECG signals, ensuring the method's clinical relevance and reliability. To reduce the impact of individual variability, the approach further incorporates crucial patient-specific information from ECG reports, such as age and gender, thus enabling accurate identification of a broad spectrum of cardiac anomalies, including rare ones. Utilizing an extensive dataset of 478,803 ECG graphic reports from real-world clinical practice, our method has demonstrated exceptional effectiveness in AD across all tested conditions, regardless of their frequency of occurrence, significantly outperforming existing models. It achieved superior performance metrics, including an AUROC of 91.2%, an F1 score of 83.7%, a sensitivity rate of 84.2%, a specificity of 83.0%, and a precision of 75.6% with a fixed recall rate of 90%. It has also demonstrated robust localization capabilities, with an AUROC of 76.5% and a Dice coefficient of 65.3% for anomaly localization.
Accelerating Distributed Deep Learning using Lossless Homomorphic Compression
Feb 12, 2024



Abstract:As deep neural networks (DNNs) grow in complexity and size, the resultant increase in communication overhead during distributed training has become a significant bottleneck, challenging the scalability of distributed training systems. Existing solutions, while aiming to mitigate this bottleneck through worker-level compression and in-network aggregation, fall short due to their inability to efficiently reconcile the trade-offs between compression effectiveness and computational overhead, hindering overall performance and scalability. In this paper, we introduce a novel compression algorithm that effectively merges worker-level compression with in-network aggregation. Our solution is both homomorphic, allowing for efficient in-network aggregation without CPU/GPU processing, and lossless, ensuring no compromise on training accuracy. Theoretically optimal in compression and computational efficiency, our approach is empirically validated across diverse DNN models such as NCF, LSTM, VGG19, and BERT-base, showing up to a 6.33$\times$ improvement in aggregation throughput and a 3.74$\times$ increase in per-iteration training speed.
Dynamic Atomic Column Detection in Transmission Electron Microscopy Videos via Ridge Estimation
Feb 02, 2023



Abstract:Ridge detection is a classical tool to extract curvilinear features in image processing. As such, it has great promise in applications to material science problems; specifically, for trend filtering relatively stable atom-shaped objects in image sequences, such as Transmission Electron Microscopy (TEM) videos. Standard analysis of TEM videos is limited to frame-by-frame object recognition. We instead harness temporal correlation across frames through simultaneous analysis of long image sequences, specified as a spatio-temporal image tensor. We define new ridge detection algorithms to non-parametrically estimate explicit trajectories of atomic-level object locations as a continuous function of time. Our approach is specially tailored to handle temporal analysis of objects that seemingly stochastically disappear and subsequently reappear throughout a sequence. We demonstrate that the proposed method is highly effective and efficient in simulation scenarios, and delivers notable performance improvements in TEM experiments compared to other material science benchmarks.
Detection and hypothesis testing of features in extremely noisy image series using topological data analysis, with applications to nanoparticle videos
Sep 28, 2022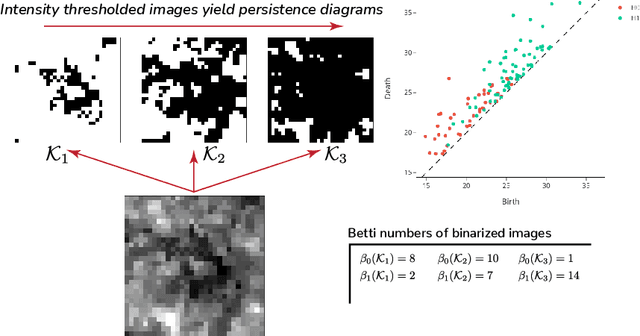


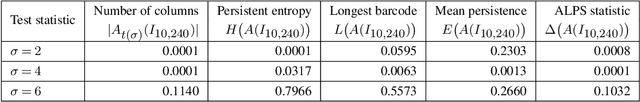
Abstract:We propose a flexible approach for the detection of features in images with ultra low signal-to-noise ratio using cubical persistent homology. Our main application is in the detection of atomic columns and other features in transmission electron microscopy (TEM) images. Cubical persistent homology is used to identify local minima in subregions in the frames of nanoparticle videos, which are hypothesized to correspond to relevant atomic features. We compare the performance of our algorithm to other employed methods for the detection of columns and their intensity. Additionally, Monte Carlo goodness-of-fit testing using real-valued summaries of persistence diagrams$\unicode{8212}$including the novel ALPS statistic$\unicode{8212}$derived from smoothed images (generated from pixels residing in the vacuum region of an image) is developed and employed to identify whether or not the proposed atomic features generated by our algorithm are due to noise. Using these summaries derived from the generated persistence diagrams, one can produce univariate time series for the nanoparticle videos, thus providing a means for assessing fluxional behavior. A guarantee on the false discovery rate for multiple Monte Carlo testing of identical hypotheses is also established.
Validation and Optimization of Multi-Organ Segmentation on Clinical Imaging Archives
Feb 10, 2020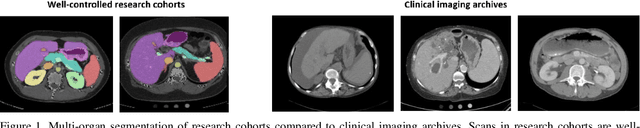
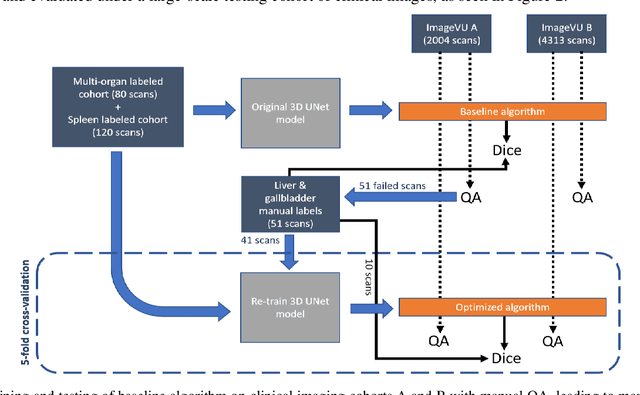
Abstract:Segmentation of abdominal computed tomography(CT) provides spatial context, morphological properties, and a framework for tissue-specific radiomics to guide quantitative Radiological assessment. A 2015 MICCAI challenge spurred substantial innovation in multi-organ abdominal CT segmentation with both traditional and deep learning methods. Recent innovations in deep methods have driven performance toward levels for which clinical translation is appealing. However, continued cross-validation on open datasets presents the risk of indirect knowledge contamination and could result in circular reasoning. Moreover, 'real world' segmentations can be challenging due to the wide variability of abdomen physiology within patients. Herein, we perform two data retrievals to capture clinically acquired deidentified abdominal CT cohorts with respect to a recently published variation on 3D U-Net (baseline algorithm). First, we retrieved 2004 deidentified studies on 476 patients with diagnosis codes involving spleen abnormalities (cohort A). Second, we retrieved 4313 deidentified studies on 1754 patients without diagnosis codes involving spleen abnormalities (cohort B). We perform prospective evaluation of the existing algorithm on both cohorts, yielding 13% and 8% failure rate, respectively. Then, we identified 51 subjects in cohort A with segmentation failures and manually corrected the liver and gallbladder labels. We re-trained the model adding the manual labels, resulting in performance improvement of 9% and 6% failure rate for the A and B cohorts, respectively. In summary, the performance of the baseline on the prospective cohorts was similar to that on previously published datasets. Moreover, adding data from the first cohort substantively improved performance when evaluated on the second withheld validation cohort.
Outlier Guided Optimization of Abdominal Segmentation
Feb 10, 2020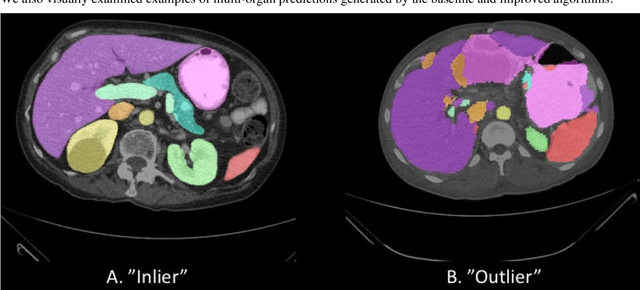
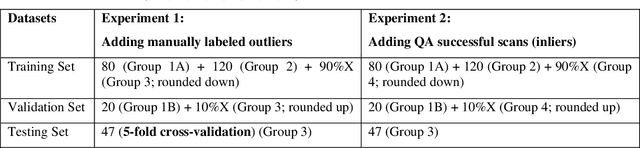
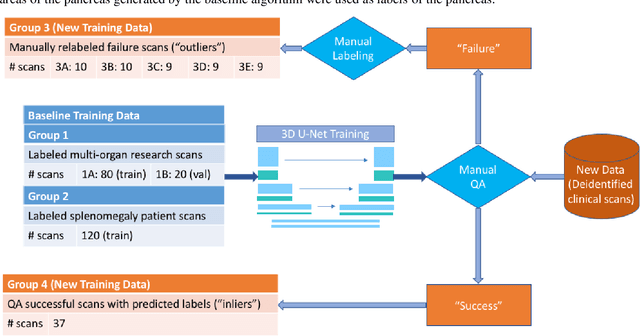
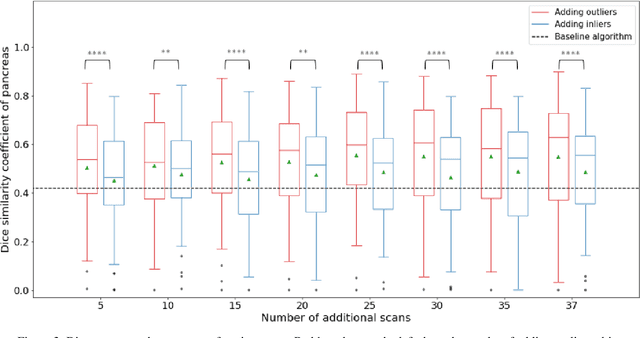
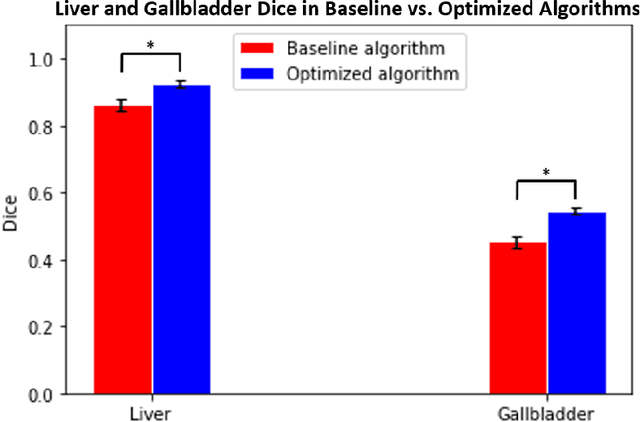
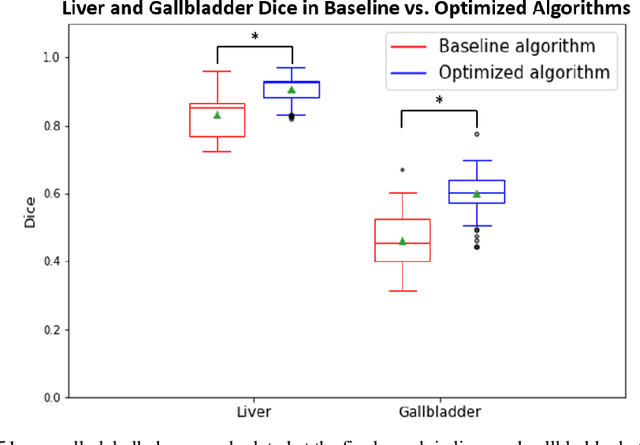
Abstract:Abdominal multi-organ segmentation of computed tomography (CT) images has been the subject of extensive research interest. It presents a substantial challenge in medical image processing, as the shape and distribution of abdominal organs can vary greatly among the population and within an individual over time. While continuous integration of novel datasets into the training set provides potential for better segmentation performance, collection of data at scale is not only costly, but also impractical in some contexts. Moreover, it remains unclear what marginal value additional data have to offer. Herein, we propose a single-pass active learning method through human quality assurance (QA). We built on a pre-trained 3D U-Net model for abdominal multi-organ segmentation and augmented the dataset either with outlier data (e.g., exemplars for which the baseline algorithm failed) or inliers (e.g., exemplars for which the baseline algorithm worked). The new models were trained using the augmented datasets with 5-fold cross-validation (for outlier data) and withheld outlier samples (for inlier data). Manual labeling of outliers increased Dice scores with outliers by 0.130, compared to an increase of 0.067 with inliers (p<0.001, two-tailed paired t-test). By adding 5 to 37 inliers or outliers to training, we find that the marginal value of adding outliers is higher than that of adding inliers. In summary, improvement on single-organ performance was obtained without diminishing multi-organ performance or significantly increasing training time. Hence, identification and correction of baseline failure cases present an effective and efficient method of selecting training data to improve algorithm performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge