Richard G. Abramson
UNesT: Local Spatial Representation Learning with Hierarchical Transformer for Efficient Medical Segmentation
Sep 28, 2022
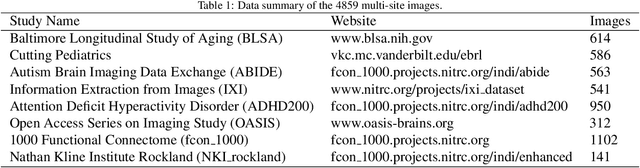


Abstract:Transformer-based models, capable of learning better global dependencies, have recently demonstrated exceptional representation learning capabilities in computer vision and medical image analysis. Transformer reformats the image into separate patches and realize global communication via the self-attention mechanism. However, positional information between patches is hard to preserve in such 1D sequences, and loss of it can lead to sub-optimal performance when dealing with large amounts of heterogeneous tissues of various sizes in 3D medical image segmentation. Additionally, current methods are not robust and efficient for heavy-duty medical segmentation tasks such as predicting a large number of tissue classes or modeling globally inter-connected tissues structures. Inspired by the nested hierarchical structures in vision transformer, we proposed a novel 3D medical image segmentation method (UNesT), employing a simplified and faster-converging transformer encoder design that achieves local communication among spatially adjacent patch sequences by aggregating them hierarchically. We extensively validate our method on multiple challenging datasets, consisting anatomies of 133 structures in brain, 14 organs in abdomen, 4 hierarchical components in kidney, and inter-connected kidney tumors). We show that UNesT consistently achieves state-of-the-art performance and evaluate its generalizability and data efficiency. Particularly, the model achieves whole brain segmentation task complete ROI with 133 tissue classes in single network, outperforms prior state-of-the-art method SLANT27 ensembled with 27 network tiles, our model performance increases the mean DSC score of the publicly available Colin and CANDI dataset from 0.7264 to 0.7444 and from 0.6968 to 0.7025, respectively.
Characterizing Renal Structures with 3D Block Aggregate Transformers
Mar 04, 2022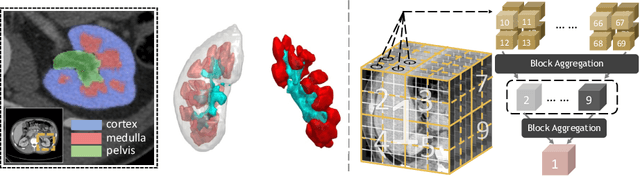
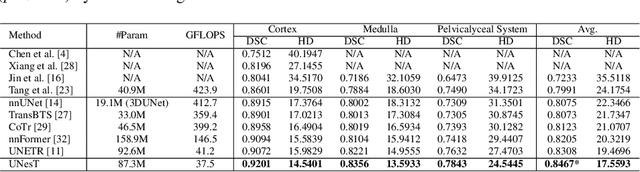
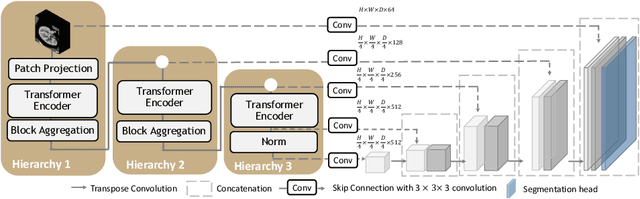
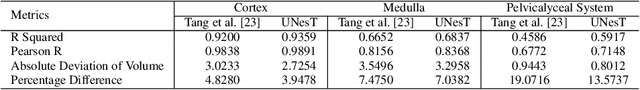
Abstract:Efficiently quantifying renal structures can provide distinct spatial context and facilitate biomarker discovery for kidney morphology. However, the development and evaluation of the transformer model to segment the renal cortex, medulla, and collecting system remains challenging due to data inefficiency. Inspired by the hierarchical structures in vision transformer, we propose a novel method using a 3D block aggregation transformer for segmenting kidney components on contrast-enhanced CT scans. We construct the first cohort of renal substructures segmentation dataset with 116 subjects under institutional review board (IRB) approval. Our method yields the state-of-the-art performance (Dice of 0.8467) against the baseline approach of 0.8308 with the data-efficient design. The Pearson R achieves 0.9891 between the proposed method and manual standards and indicates the strong correlation and reproducibility for volumetric analysis. We extend the proposed method to the public KiTS dataset, the method leads to improved accuracy compared to transformer-based approaches. We show that the 3D block aggregation transformer can achieve local communication between sequence representations without modifying self-attention, and it can serve as an accurate and efficient quantification tool for characterizing renal structures.
RAP-Net: Coarse-to-Fine Multi-Organ Segmentation with Single Random Anatomical Prior
Dec 24, 2020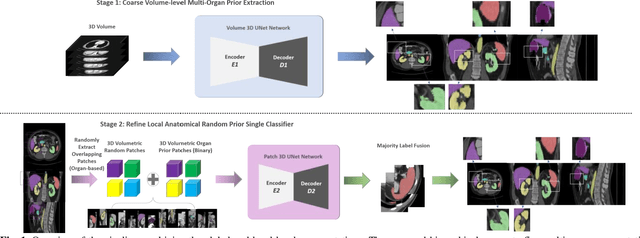
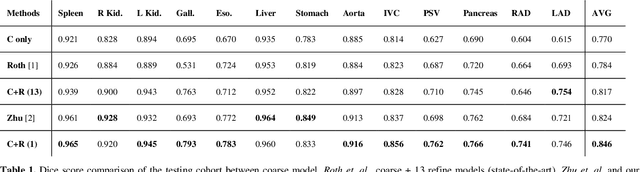
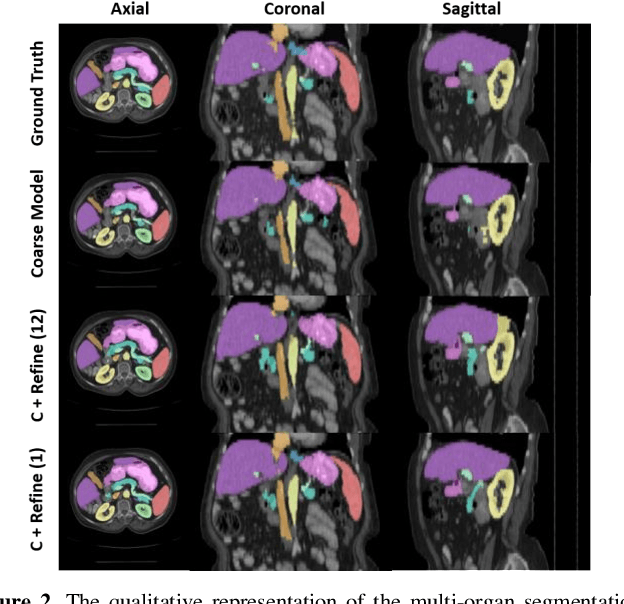
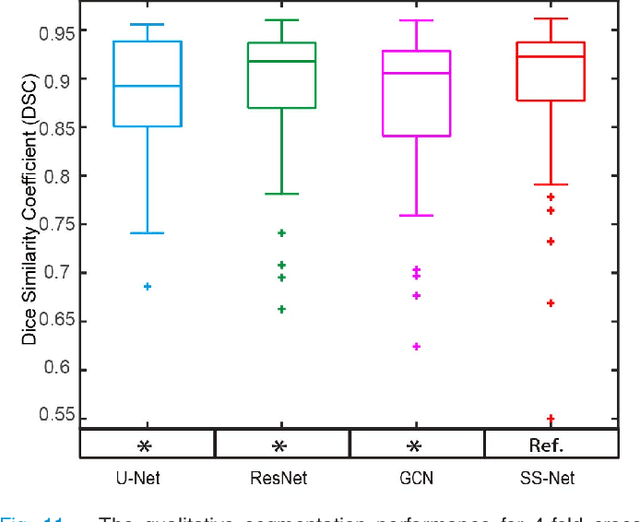
Abstract:Performing coarse-to-fine abdominal multi-organ segmentation facilitates to extract high-resolution segmentation minimizing the lost of spatial contextual information. However, current coarse-to-refine approaches require a significant number of models to perform single organ refine segmentation corresponding to the extracted organ region of interest (ROI). We propose a coarse-to-fine pipeline, which starts from the extraction of the global prior context of multiple organs from 3D volumes using a low-resolution coarse network, followed by a fine phase that uses a single refined model to segment all abdominal organs instead of multiple organ corresponding models. We combine the anatomical prior with corresponding extracted patches to preserve the anatomical locations and boundary information for performing high-resolution segmentation across all organs in a single model. To train and evaluate our method, a clinical research cohort consisting of 100 patient volumes with 13 organs well-annotated is used. We tested our algorithms with 4-fold cross-validation and computed the Dice score for evaluating the segmentation performance of the 13 organs. Our proposed method using single auto-context outperforms the state-of-the-art on 13 models with an average Dice score 84.58% versus 81.69% (p<0.0001).
Validation and Optimization of Multi-Organ Segmentation on Clinical Imaging Archives
Feb 10, 2020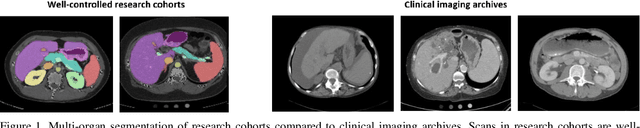
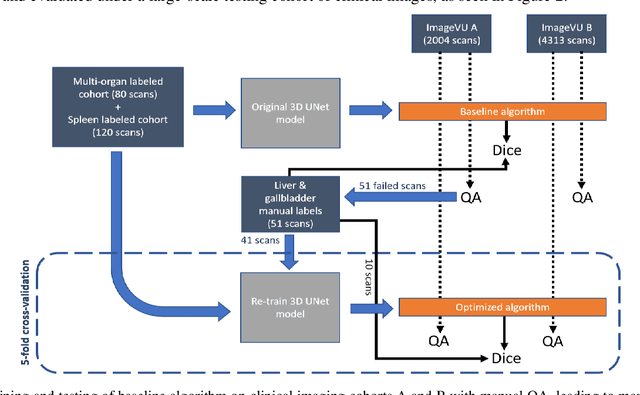
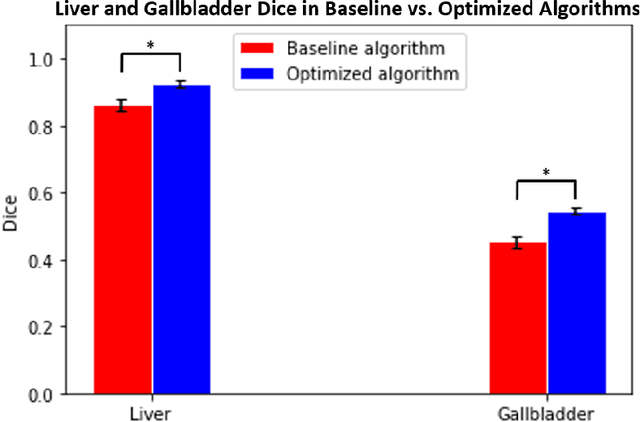
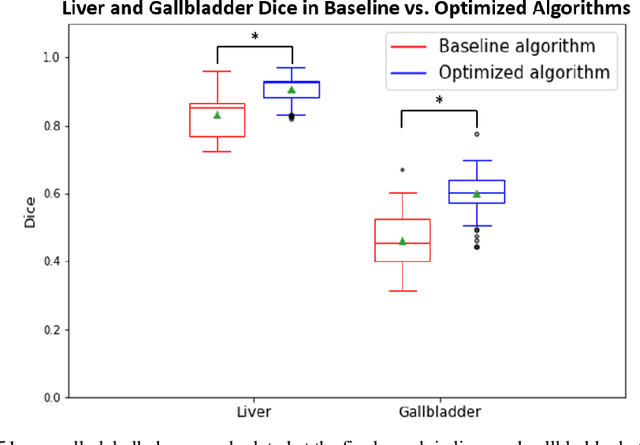
Abstract:Segmentation of abdominal computed tomography(CT) provides spatial context, morphological properties, and a framework for tissue-specific radiomics to guide quantitative Radiological assessment. A 2015 MICCAI challenge spurred substantial innovation in multi-organ abdominal CT segmentation with both traditional and deep learning methods. Recent innovations in deep methods have driven performance toward levels for which clinical translation is appealing. However, continued cross-validation on open datasets presents the risk of indirect knowledge contamination and could result in circular reasoning. Moreover, 'real world' segmentations can be challenging due to the wide variability of abdomen physiology within patients. Herein, we perform two data retrievals to capture clinically acquired deidentified abdominal CT cohorts with respect to a recently published variation on 3D U-Net (baseline algorithm). First, we retrieved 2004 deidentified studies on 476 patients with diagnosis codes involving spleen abnormalities (cohort A). Second, we retrieved 4313 deidentified studies on 1754 patients without diagnosis codes involving spleen abnormalities (cohort B). We perform prospective evaluation of the existing algorithm on both cohorts, yielding 13% and 8% failure rate, respectively. Then, we identified 51 subjects in cohort A with segmentation failures and manually corrected the liver and gallbladder labels. We re-trained the model adding the manual labels, resulting in performance improvement of 9% and 6% failure rate for the A and B cohorts, respectively. In summary, the performance of the baseline on the prospective cohorts was similar to that on previously published datasets. Moreover, adding data from the first cohort substantively improved performance when evaluated on the second withheld validation cohort.
Outlier Guided Optimization of Abdominal Segmentation
Feb 10, 2020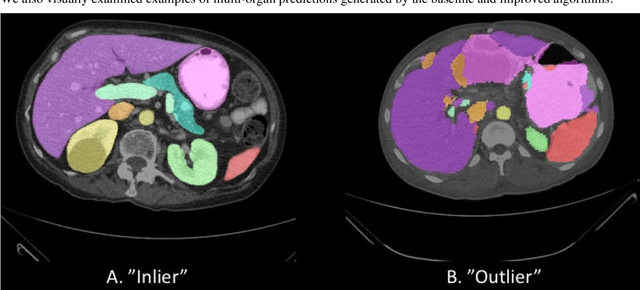
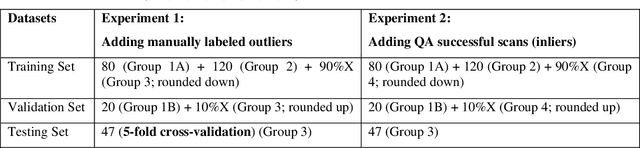
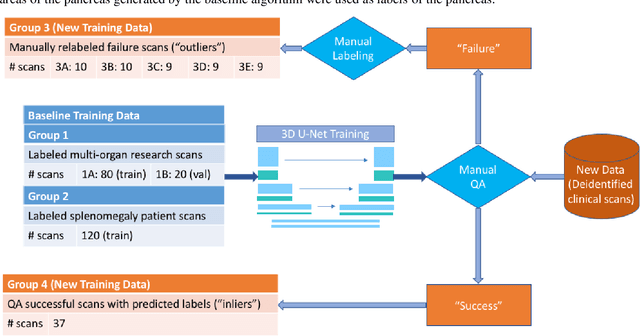
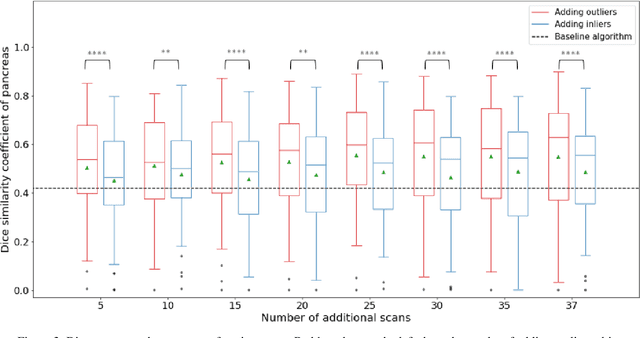
Abstract:Abdominal multi-organ segmentation of computed tomography (CT) images has been the subject of extensive research interest. It presents a substantial challenge in medical image processing, as the shape and distribution of abdominal organs can vary greatly among the population and within an individual over time. While continuous integration of novel datasets into the training set provides potential for better segmentation performance, collection of data at scale is not only costly, but also impractical in some contexts. Moreover, it remains unclear what marginal value additional data have to offer. Herein, we propose a single-pass active learning method through human quality assurance (QA). We built on a pre-trained 3D U-Net model for abdominal multi-organ segmentation and augmented the dataset either with outlier data (e.g., exemplars for which the baseline algorithm failed) or inliers (e.g., exemplars for which the baseline algorithm worked). The new models were trained using the augmented datasets with 5-fold cross-validation (for outlier data) and withheld outlier samples (for inlier data). Manual labeling of outliers increased Dice scores with outliers by 0.130, compared to an increase of 0.067 with inliers (p<0.001, two-tailed paired t-test). By adding 5 to 37 inliers or outliers to training, we find that the marginal value of adding outliers is higher than that of adding inliers. In summary, improvement on single-organ performance was obtained without diminishing multi-organ performance or significantly increasing training time. Hence, identification and correction of baseline failure cases present an effective and efficient method of selecting training data to improve algorithm performance.
Stochastic tissue window normalization of deep learning on computed tomography
Dec 01, 2019


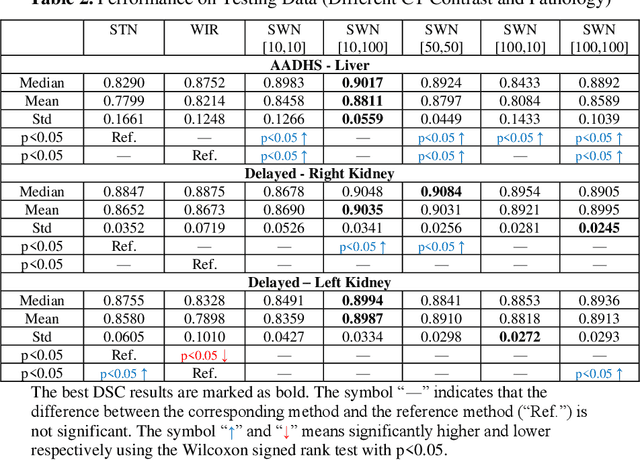
Abstract:Tissue window filtering has been widely used in deep learning for computed tomography (CT) image analyses to improve training performance (e.g., soft tissue windows for abdominal CT). However, the effectiveness of tissue window normalization is questionable since the generalizability of the trained model might be further harmed, especially when such models are applied to new cohorts with different CT reconstruction kernels, contrast mechanisms, dynamic variations in the acquisition, and physiological changes. We evaluate the effectiveness of both with and without using soft tissue window normalization on multisite CT cohorts. Moreover, we propose a stochastic tissue window normalization (SWN) method to improve the generalizability of tissue window normalization. Different from the random sampling, the SWN method centers the randomization around the soft tissue window to maintain the specificity for abdominal organs. To evaluate the performance of different strategies, 80 training and 453 validation and testing scans from six datasets are employed to perform multi-organ segmentation using standard 2D U-Net. The six datasets cover the scenarios, where the training and testing scans are from (1) same scanner and same population, (2) same CT contrast but different pathology, and (3) different CT contrast and pathology. The traditional soft tissue window and nonwindowed approaches achieved better performance on (1). The proposed SWN achieved general superior performance on (2) and (3) with statistical analyses, which offers better generalizability for a trained model.
Contrast Phase Classification with a Generative Adversarial Network
Nov 14, 2019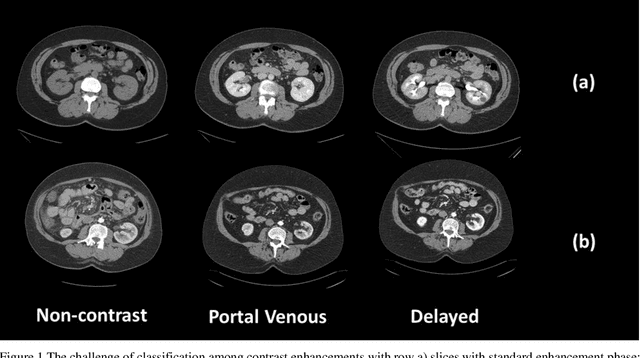
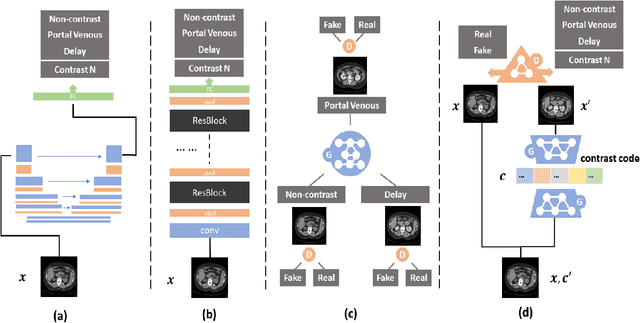
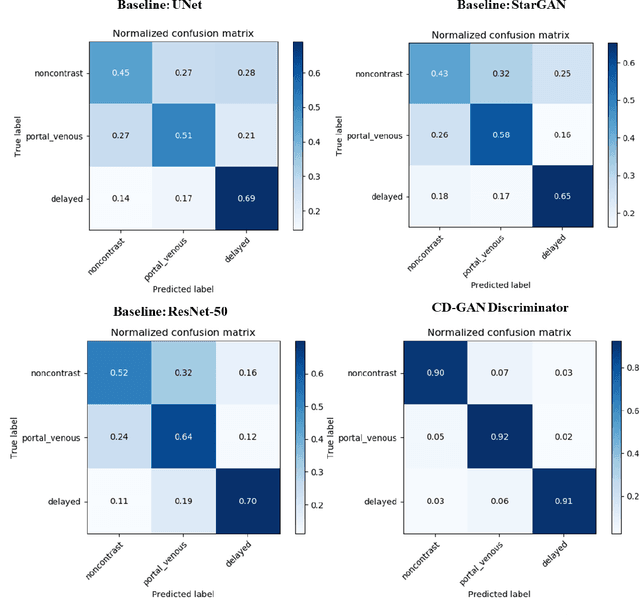
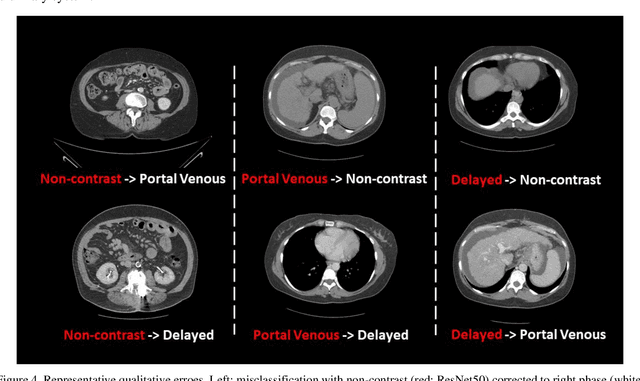
Abstract:Dynamic contrast enhanced computed tomography (CT) is an imaging technique that provides critical information on the relationship of vascular structure and dynamics in the context of underlying anatomy. A key challenge for image processing with contrast enhanced CT is that phase discrepancies are latent in different tissues due to contrast protocols, vascular dynamics, and metabolism variance. Previous studies with deep learning frameworks have been proposed for classifying contrast enhancement with networks inspired by computer vision. Here, we revisit the challenge in the context of whole abdomen contrast enhanced CTs. To capture and compensate for the complex contrast changes, we propose a novel discriminator in the form of a multi-domain disentangled representation learning network. The goal of this network is to learn an intermediate representation that separates contrast enhancement from anatomy and enables classification of images with varying contrast time. Briefly, our unpaired contrast disentangling GAN(CD-GAN) Discriminator follows the ResNet architecture to classify a CT scan from different enhancement phases. To evaluate the approach, we trained the enhancement phase classifier on 21060 slices from two clinical cohorts of 230 subjects. Testing was performed on 9100 slices from 30 independent subjects who had been imaged with CT scans from all contrast phases. Performance was quantified in terms of the multi-class normalized confusion matrix. The proposed network significantly improved correspondence over baseline UNet, ResNet50 and StarGAN performance of accuracy scores 0.54. 0.55, 0.62 and 0.91, respectively. The proposed discriminator from the disentangled network presents a promising technique that may allow deeper modeling of dynamic imaging against patient specific anatomies.
* 8 pages, 4 figures
Semi-Supervised Multi-Organ Segmentation through Quality Assurance Supervision
Nov 12, 2019



Abstract:Human in-the-loop quality assurance (QA) is typically performed after medical image segmentation to ensure that the systems are performing as intended, as well as identifying and excluding outliers. By performing QA on large-scale, previously unlabeled testing data, categorical QA scores can be generatedIn this paper, we propose a semi-supervised multi-organ segmentation deep neural network consisting of a traditional segmentation model generator and a QA involved discriminator. A large-scale dataset of 2027 volumes are used to train the generator, whose 2-D montage images and segmentation mask with QA scores are used to train the discriminator. To generate the QA scores, the 2-D montage images were reviewed manually and coded 0 (success), 1 (errors consistent with published performance), and 2 (gross failure). Then, the ResNet-18 network was trained with 1623 montage images in equal distribution of all three code labels and achieved an accuracy 94% for classification predictions with 404 montage images withheld for the test cohort. To assess the performance of using the QA supervision, the discriminator was used as a loss function in a multi-organ segmentation pipeline. The inclusion of QA-loss function boosted performance on the unlabeled test dataset from 714 patients to 951 patients over the baseline model. Additionally, the number of failures decreased from 606 (29.90%) to 402 (19.83%). The contributions of the proposed method are threefold: We show that (1) the QA scores can be used as a loss function to perform semi-supervised learning for unlabeled data, (2) the well trained discriminator is learnt by QA score rather than traditional true/false, and (3) the performance of multi-organ segmentation on unlabeled datasets can be fine-tuned with more robust and higher accuracy than the original baseline method.
Splenomegaly Segmentation on Multi-modal MRI using Deep Convolutional Networks
Nov 09, 2018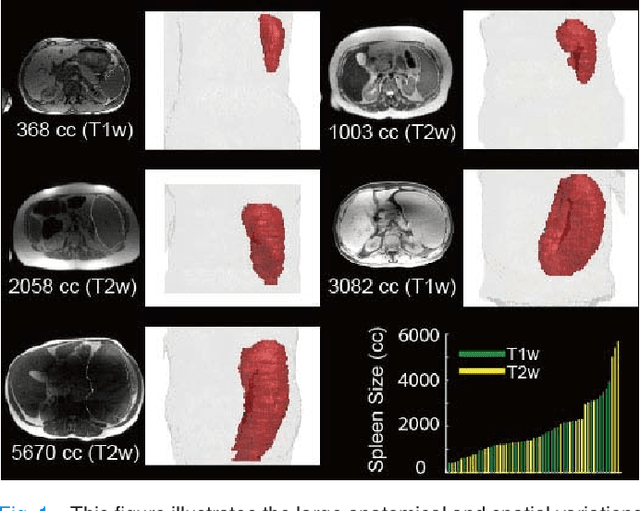
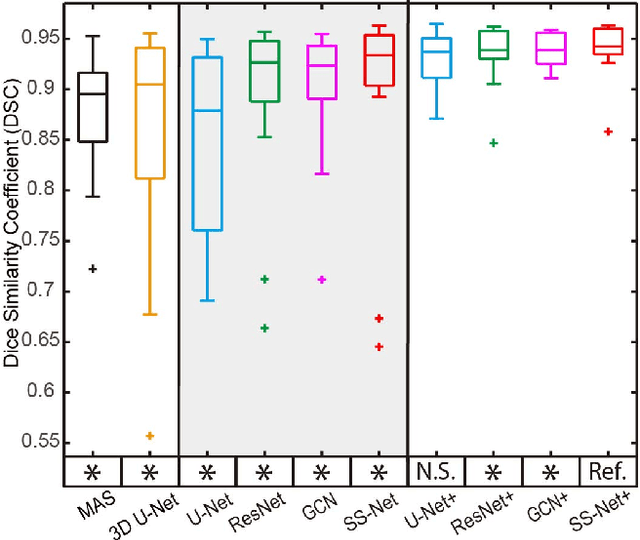
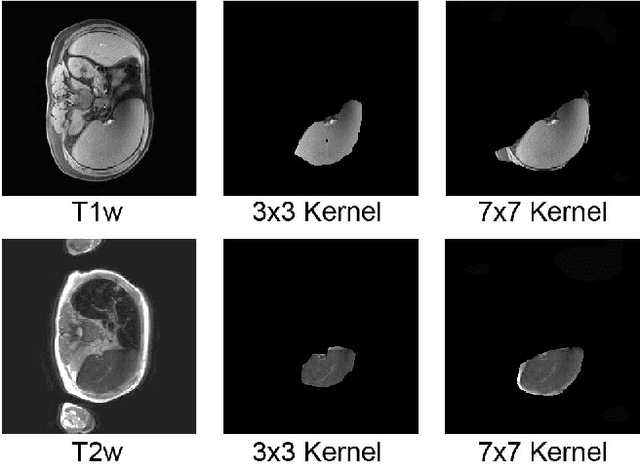
Abstract:The findings of splenomegaly, abnormal enlargement of the spleen, is a non-invasive clinical biomarker for liver and spleen disease. Automated segmentation methods are essential to efficiently quantify splenomegaly from clinically acquired abdominal magnetic resonance imaging (MRI) scans. However, the task is challenging due to (1) large anatomical and spatial variations of splenomegaly, (2) large inter- and intra-scan intensity variations on multi-modal MRI, and (3) limited numbers of labeled splenomegaly scans. In this paper, we propose the Splenomegaly Segmentation Network (SS-Net) to introduce the deep convolutional neural network (DCNN) approaches in multi-modal MRI splenomegaly segmentation. Large convolutional kernel layers were used to address the spatial and anatomical variations, while the conditional generative adversarial networks (GAN) were employed to leverage the segmentation performance of SS-Net in an end-to-end manner. A clinically acquired cohort containing both T1-weighted (T1w) and T2-weighted (T2w) MRI splenomegaly scans was used to train and evaluate the performance of multi-atlas segmentation (MAS), 2D DCNN networks, and a 3D DCNN network. From the experimental results, the DCNN methods achieved superior performance to the state-of-the-art MAS method. The proposed SS-Net method achieved the highest median and mean Dice scores among investigated baseline DCNN methods.
SynSeg-Net: Synthetic Segmentation Without Target Modality Ground Truth
Oct 15, 2018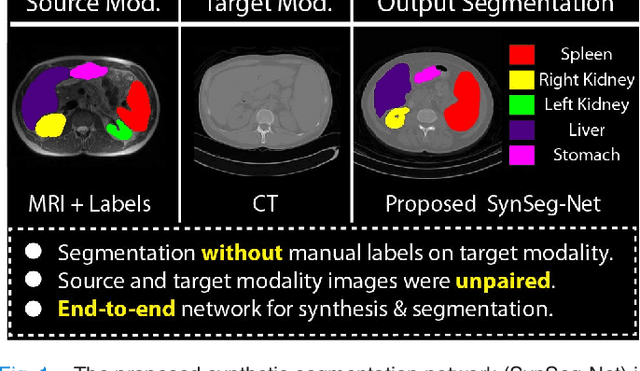
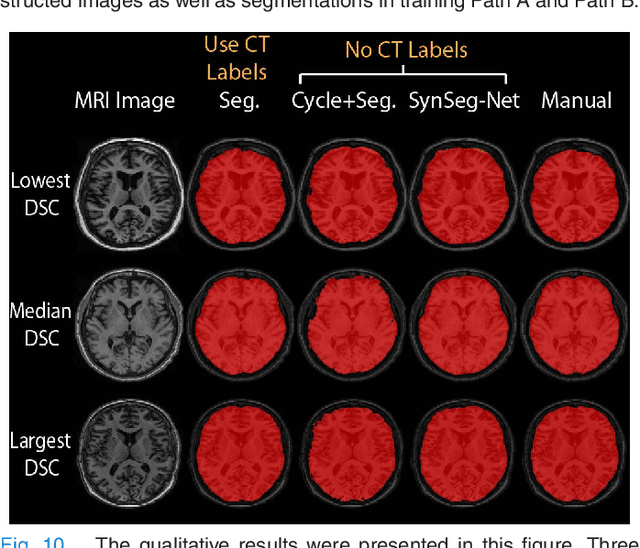
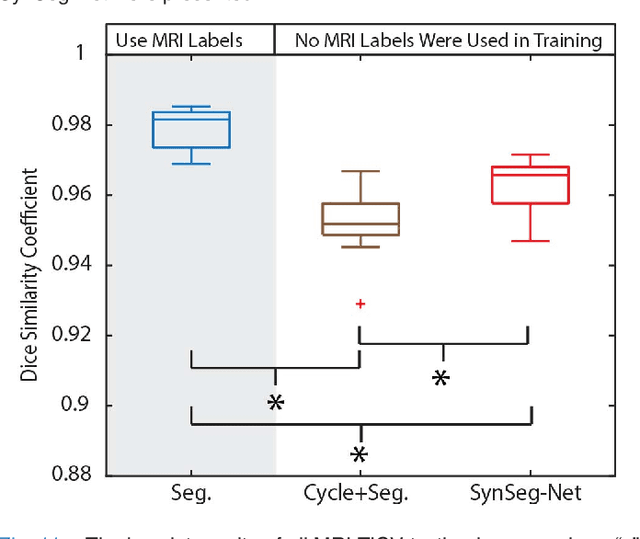
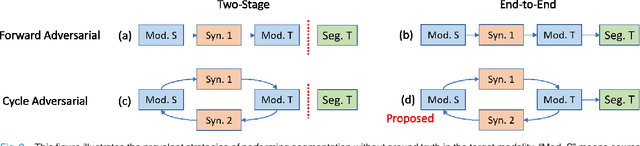
Abstract:A key limitation of deep convolutional neural networks (DCNN) based image segmentation methods is the lack of generalizability. Manually traced training images are typically required when segmenting organs in a new imaging modality or from distinct disease cohort. The manual efforts can be alleviated if the manually traced images in one imaging modality (e.g., MRI) are able to train a segmentation network for another imaging modality (e.g., CT). In this paper, we propose an end-to-end synthetic segmentation network (SynSeg-Net) to train a segmentation network for a target imaging modality without having manual labels. SynSeg-Net is trained by using (1) unpaired intensity images from source and target modalities, and (2) manual labels only from source modality. SynSeg-Net is enabled by the recent advances of cycle generative adversarial networks (CycleGAN) and DCNN. We evaluate the performance of the SynSeg-Net on two experiments: (1) MRI to CT splenomegaly synthetic segmentation for abdominal images, and (2) CT to MRI total intracranial volume synthetic segmentation (TICV) for brain images. The proposed end-to-end approach achieved superior performance to two stage methods. Moreover, the SynSeg-Net achieved comparable performance to the traditional segmentation network using target modality labels in certain scenarios. The source code of SynSeg-Net is publicly available (https://github.com/MASILab/SynSeg-Net).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge