Characterizing Renal Structures with 3D Block Aggregate Transformers
Paper and Code
Mar 04, 2022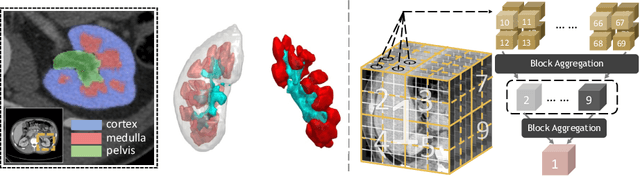
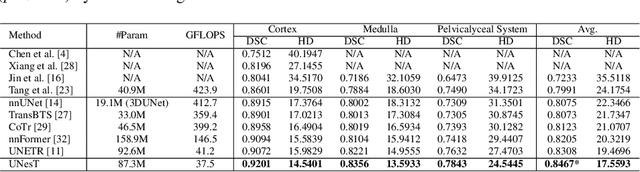
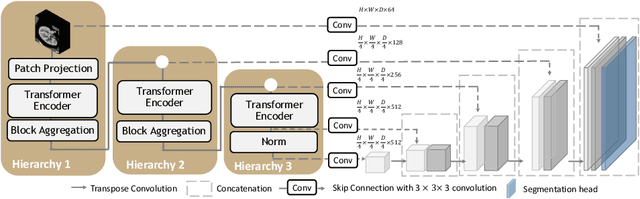
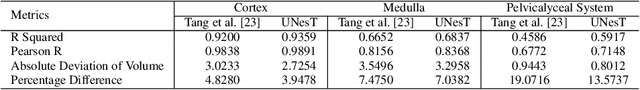
Efficiently quantifying renal structures can provide distinct spatial context and facilitate biomarker discovery for kidney morphology. However, the development and evaluation of the transformer model to segment the renal cortex, medulla, and collecting system remains challenging due to data inefficiency. Inspired by the hierarchical structures in vision transformer, we propose a novel method using a 3D block aggregation transformer for segmenting kidney components on contrast-enhanced CT scans. We construct the first cohort of renal substructures segmentation dataset with 116 subjects under institutional review board (IRB) approval. Our method yields the state-of-the-art performance (Dice of 0.8467) against the baseline approach of 0.8308 with the data-efficient design. The Pearson R achieves 0.9891 between the proposed method and manual standards and indicates the strong correlation and reproducibility for volumetric analysis. We extend the proposed method to the public KiTS dataset, the method leads to improved accuracy compared to transformer-based approaches. We show that the 3D block aggregation transformer can achieve local communication between sequence representations without modifying self-attention, and it can serve as an accurate and efficient quantification tool for characterizing renal structures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge