Deep Gandhi
RobustDebias: Debiasing Language Models using Distributionally Robust Optimization
Jan 30, 2026Abstract:Pretrained language models have been shown to exhibit biases and social stereotypes. Prior work on debiasing these models has largely focused on modifying embedding spaces during pretraining, which is not scalable for large models. Fine-tuning pretrained models on task-specific datasets can both degrade model performance and amplify biases present in the fine-tuning data. We address bias amplification during fine-tuning rather than costly pretraining, focusing on BERT models due to their widespread use in language understanding tasks. While Empirical Risk Minimization effectively optimizes downstream performance, it often amplifies social biases during fine-tuning. To counter this, we propose \textit{RobustDebias}, a novel mechanism which adapts Distributionally Robust Optimization (DRO) to debias language models during fine-tuning. Our approach debiases models across multiple demographics during MLM fine-tuning and generalizes to any dataset or task. Extensive experiments on various language models show significant bias mitigation with minimal performance impact.
BraTS-PEDs: Results of the Multi-Consortium International Pediatric Brain Tumor Segmentation Challenge 2023
Jul 11, 2024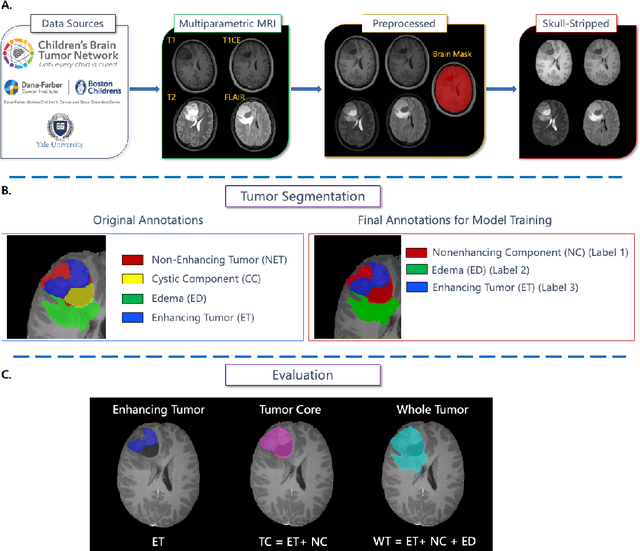

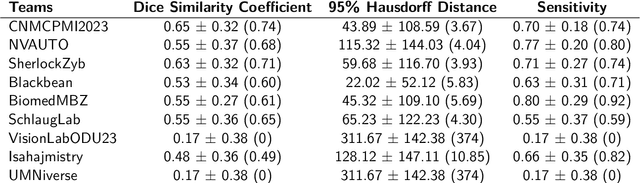
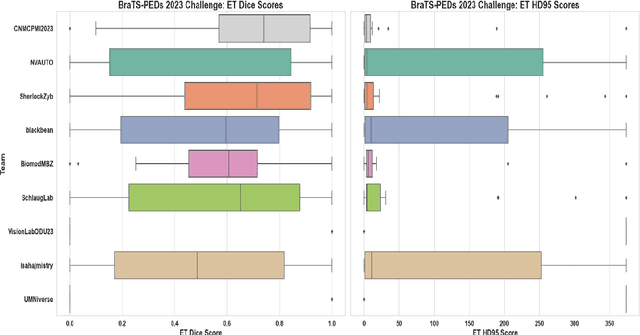
Abstract:Pediatric central nervous system tumors are the leading cause of cancer-related deaths in children. The five-year survival rate for high-grade glioma in children is less than 20%. The development of new treatments is dependent upon multi-institutional collaborative clinical trials requiring reproducible and accurate centralized response assessment. We present the results of the BraTS-PEDs 2023 challenge, the first Brain Tumor Segmentation (BraTS) challenge focused on pediatric brain tumors. This challenge utilized data acquired from multiple international consortia dedicated to pediatric neuro-oncology and clinical trials. BraTS-PEDs 2023 aimed to evaluate volumetric segmentation algorithms for pediatric brain gliomas from magnetic resonance imaging using standardized quantitative performance evaluation metrics employed across the BraTS 2023 challenges. The top-performing AI approaches for pediatric tumor analysis included ensembles of nnU-Net and Swin UNETR, Auto3DSeg, or nnU-Net with a self-supervised framework. The BraTSPEDs 2023 challenge fostered collaboration between clinicians (neuro-oncologists, neuroradiologists) and AI/imaging scientists, promoting faster data sharing and the development of automated volumetric analysis techniques. These advancements could significantly benefit clinical trials and improve the care of children with brain tumors.
The Brain Tumor Segmentation in Pediatrics (BraTS-PEDs) Challenge: Focus on Pediatrics (CBTN-CONNECT-DIPGR-ASNR-MICCAI BraTS-PEDs)
Apr 29, 2024Abstract:Pediatric tumors of the central nervous system are the most common cause of cancer-related death in children. The five-year survival rate for high-grade gliomas in children is less than 20%. Due to their rarity, the diagnosis of these entities is often delayed, their treatment is mainly based on historic treatment concepts, and clinical trials require multi-institutional collaborations. Here we present the CBTN-CONNECT-DIPGR-ASNR-MICCAI BraTS-PEDs challenge, focused on pediatric brain tumors with data acquired across multiple international consortia dedicated to pediatric neuro-oncology and clinical trials. The CBTN-CONNECT-DIPGR-ASNR-MICCAI BraTS-PEDs challenge brings together clinicians and AI/imaging scientists to lead to faster development of automated segmentation techniques that could benefit clinical trials, and ultimately the care of children with brain tumors.
Training and Comparison of nnU-Net and DeepMedic Methods for Autosegmentation of Pediatric Brain Tumors
Jan 30, 2024



Abstract:Brain tumors are the most common solid tumors and the leading cause of cancer-related death among children. Tumor segmentation is essential in surgical and treatment planning, and response assessment and monitoring. However, manual segmentation is time-consuming and has high inter-operator variability, underscoring the need for more efficient methods. We compared two deep learning-based 3D segmentation models, DeepMedic and nnU-Net, after training with pediatric-specific multi-institutional brain tumor data using based on multi-parametric MRI scans.Multi-parametric preoperative MRI scans of 339 pediatric patients (n=293 internal and n=46 external cohorts) with a variety of tumor subtypes, were preprocessed and manually segmented into four tumor subregions, i.e., enhancing tumor (ET), non-enhancing tumor (NET), cystic components (CC), and peritumoral edema (ED). After training, performance of the two models on internal and external test sets was evaluated using Dice scores, sensitivity, and Hausdorff distance with reference to ground truth manual segmentations. Dice score for nnU-Net internal test sets was (mean +/- SD (median)) 0.9+/-0.07 (0.94) for WT, 0.77+/-0.29 for ET, 0.66+/-0.32 for NET, 0.71+/-0.33 for CC, and 0.71+/-0.40 for ED, respectively. For DeepMedic the Dice scores were 0.82+/-0.16 for WT, 0.66+/-0.32 for ET, 0.48+/-0.27, for NET, 0.48+/-0.36 for CC, and 0.19+/-0.33 for ED, respectively. Dice scores were significantly higher for nnU-Net (p<=0.01). External validation of the trained nnU-Net model on the multi-institutional BraTS-PEDs 2023 dataset revealed high generalization capability in segmentation of whole tumor and tumor core with Dice scores of 0.87+/-0.13 (0.91) and 0.83+/-0.18 (0.89), respectively. Pediatric-specific data trained nnU-Net model is superior to DeepMedic for whole tumor and subregion segmentation of pediatric brain tumors.
A Federated Approach for Hate Speech Detection
Feb 18, 2023



Abstract:Hate speech detection has been the subject of high research attention, due to the scale of content created on social media. In spite of the attention and the sensitive nature of the task, privacy preservation in hate speech detection has remained under-studied. The majority of research has focused on centralised machine learning infrastructures which risk leaking data. In this paper, we show that using federated machine learning can help address privacy the concerns that are inherent to hate speech detection while obtaining up to 6.81% improvement in terms of F1-score.
A Federated Approach to Predicting Emojis in Hindi Tweets
Nov 11, 2022Abstract:The use of emojis affords a visual modality to, often private, textual communication. The task of predicting emojis however provides a challenge for machine learning as emoji use tends to cluster into the frequently used and the rarely used emojis. Much of the machine learning research on emoji use has focused on high resource languages and has conceptualised the task of predicting emojis around traditional server-side machine learning approaches. However, traditional machine learning approaches for private communication can introduce privacy concerns, as these approaches require all data to be transmitted to a central storage. In this paper, we seek to address the dual concerns of emphasising high resource languages for emoji prediction and risking the privacy of people's data. We introduce a new dataset of $118$k tweets (augmented from $25$k unique tweets) for emoji prediction in Hindi, and propose a modification to the federated learning algorithm, CausalFedGSD, which aims to strike a balance between model performance and user privacy. We show that our approach obtains comparative scores with more complex centralised models while reducing the amount of data required to optimise the models and minimising risks to user privacy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge