Debanjan Haldar
BraTS-PEDs: Results of the Multi-Consortium International Pediatric Brain Tumor Segmentation Challenge 2023
Jul 11, 2024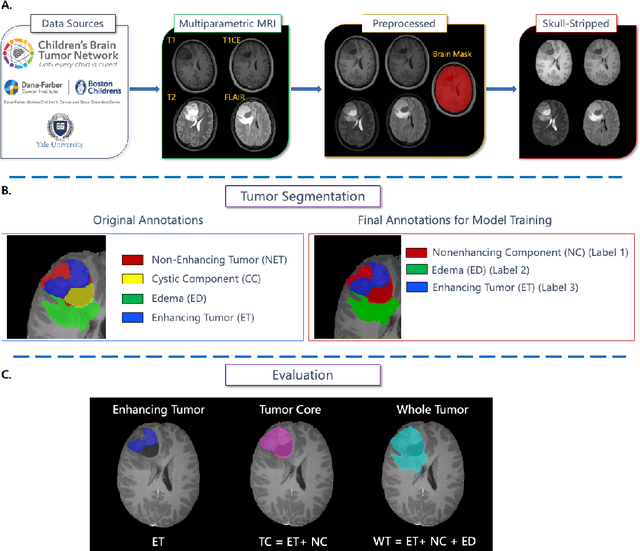

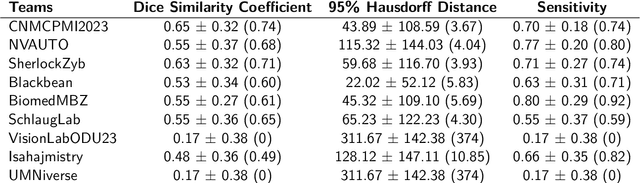
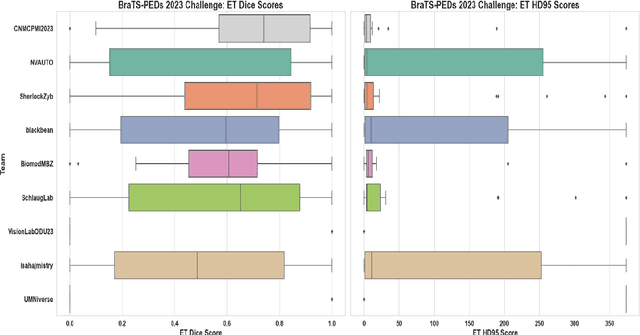
Abstract:Pediatric central nervous system tumors are the leading cause of cancer-related deaths in children. The five-year survival rate for high-grade glioma in children is less than 20%. The development of new treatments is dependent upon multi-institutional collaborative clinical trials requiring reproducible and accurate centralized response assessment. We present the results of the BraTS-PEDs 2023 challenge, the first Brain Tumor Segmentation (BraTS) challenge focused on pediatric brain tumors. This challenge utilized data acquired from multiple international consortia dedicated to pediatric neuro-oncology and clinical trials. BraTS-PEDs 2023 aimed to evaluate volumetric segmentation algorithms for pediatric brain gliomas from magnetic resonance imaging using standardized quantitative performance evaluation metrics employed across the BraTS 2023 challenges. The top-performing AI approaches for pediatric tumor analysis included ensembles of nnU-Net and Swin UNETR, Auto3DSeg, or nnU-Net with a self-supervised framework. The BraTSPEDs 2023 challenge fostered collaboration between clinicians (neuro-oncologists, neuroradiologists) and AI/imaging scientists, promoting faster data sharing and the development of automated volumetric analysis techniques. These advancements could significantly benefit clinical trials and improve the care of children with brain tumors.
The Brain Tumor Segmentation in Pediatrics (BraTS-PEDs) Challenge: Focus on Pediatrics (CBTN-CONNECT-DIPGR-ASNR-MICCAI BraTS-PEDs)
Apr 29, 2024Abstract:Pediatric tumors of the central nervous system are the most common cause of cancer-related death in children. The five-year survival rate for high-grade gliomas in children is less than 20%. Due to their rarity, the diagnosis of these entities is often delayed, their treatment is mainly based on historic treatment concepts, and clinical trials require multi-institutional collaborations. Here we present the CBTN-CONNECT-DIPGR-ASNR-MICCAI BraTS-PEDs challenge, focused on pediatric brain tumors with data acquired across multiple international consortia dedicated to pediatric neuro-oncology and clinical trials. The CBTN-CONNECT-DIPGR-ASNR-MICCAI BraTS-PEDs challenge brings together clinicians and AI/imaging scientists to lead to faster development of automated segmentation techniques that could benefit clinical trials, and ultimately the care of children with brain tumors.
Training and Comparison of nnU-Net and DeepMedic Methods for Autosegmentation of Pediatric Brain Tumors
Jan 30, 2024



Abstract:Brain tumors are the most common solid tumors and the leading cause of cancer-related death among children. Tumor segmentation is essential in surgical and treatment planning, and response assessment and monitoring. However, manual segmentation is time-consuming and has high inter-operator variability, underscoring the need for more efficient methods. We compared two deep learning-based 3D segmentation models, DeepMedic and nnU-Net, after training with pediatric-specific multi-institutional brain tumor data using based on multi-parametric MRI scans.Multi-parametric preoperative MRI scans of 339 pediatric patients (n=293 internal and n=46 external cohorts) with a variety of tumor subtypes, were preprocessed and manually segmented into four tumor subregions, i.e., enhancing tumor (ET), non-enhancing tumor (NET), cystic components (CC), and peritumoral edema (ED). After training, performance of the two models on internal and external test sets was evaluated using Dice scores, sensitivity, and Hausdorff distance with reference to ground truth manual segmentations. Dice score for nnU-Net internal test sets was (mean +/- SD (median)) 0.9+/-0.07 (0.94) for WT, 0.77+/-0.29 for ET, 0.66+/-0.32 for NET, 0.71+/-0.33 for CC, and 0.71+/-0.40 for ED, respectively. For DeepMedic the Dice scores were 0.82+/-0.16 for WT, 0.66+/-0.32 for ET, 0.48+/-0.27, for NET, 0.48+/-0.36 for CC, and 0.19+/-0.33 for ED, respectively. Dice scores were significantly higher for nnU-Net (p<=0.01). External validation of the trained nnU-Net model on the multi-institutional BraTS-PEDs 2023 dataset revealed high generalization capability in segmentation of whole tumor and tumor core with Dice scores of 0.87+/-0.13 (0.91) and 0.83+/-0.18 (0.89), respectively. Pediatric-specific data trained nnU-Net model is superior to DeepMedic for whole tumor and subregion segmentation of pediatric brain tumors.
A multi-institutional pediatric dataset of clinical radiology MRIs by the Children's Brain Tumor Network
Oct 02, 2023

Abstract:Pediatric brain and spinal cancers remain the leading cause of cancer-related death in children. Advancements in clinical decision-support in pediatric neuro-oncology utilizing the wealth of radiology imaging data collected through standard care, however, has significantly lagged other domains. Such data is ripe for use with predictive analytics such as artificial intelligence (AI) methods, which require large datasets. To address this unmet need, we provide a multi-institutional, large-scale pediatric dataset of 23,101 multi-parametric MRI exams acquired through routine care for 1,526 brain tumor patients, as part of the Children's Brain Tumor Network. This includes longitudinal MRIs across various cancer diagnoses, with associated patient-level clinical information, digital pathology slides, as well as tissue genotype and omics data. To facilitate downstream analysis, treatment-na\"ive images for 370 subjects were processed and released through the NCI Childhood Cancer Data Initiative via the Cancer Data Service. Through ongoing efforts to continuously build these imaging repositories, our aim is to accelerate discovery and translational AI models with real-world data, to ultimately empower precision medicine for children.
The Brain Tumor Segmentation Challenge 2023: Focus on Pediatrics
May 26, 2023Abstract:Pediatric tumors of the central nervous system are the most common cause of cancer-related death in children. The five-year survival rate for high-grade gliomas in children is less than 20\%. Due to their rarity, the diagnosis of these entities is often delayed, their treatment is mainly based on historic treatment concepts, and clinical trials require multi-institutional collaborations. The MICCAI Brain Tumor Segmentation (BraTS) Challenge is a landmark community benchmark event with a successful history of 12 years of resource creation for the segmentation and analysis of adult glioma. Here we present the CBTN-CONNECT-DIPGR-ASNR-MICCAI BraTS-PEDs 2023 challenge, which represents the first BraTS challenge focused on pediatric brain tumors with data acquired across multiple international consortia dedicated to pediatric neuro-oncology and clinical trials. The BraTS-PEDs 2023 challenge focuses on benchmarking the development of volumentric segmentation algorithms for pediatric brain glioma through standardized quantitative performance evaluation metrics utilized across the BraTS 2023 cluster of challenges. Models gaining knowledge from the BraTS-PEDs multi-parametric structural MRI (mpMRI) training data will be evaluated on separate validation and unseen test mpMRI dataof high-grade pediatric glioma. The CBTN-CONNECT-DIPGR-ASNR-MICCAI BraTS-PEDs 2023 challenge brings together clinicians and AI/imaging scientists to lead to faster development of automated segmentation techniques that could benefit clinical trials, and ultimately the care of children with brain tumors.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge