Matthew S. Rosen
From Low Field to High Value: Robust Cortical Mapping from Low-Field MRI
May 18, 2025Abstract:Three-dimensional reconstruction of cortical surfaces from MRI for morphometric analysis is fundamental for understanding brain structure. While high-field MRI (HF-MRI) is standard in research and clinical settings, its limited availability hinders widespread use. Low-field MRI (LF-MRI), particularly portable systems, offers a cost-effective and accessible alternative. However, existing cortical surface analysis tools are optimized for high-resolution HF-MRI and struggle with the lower signal-to-noise ratio and resolution of LF-MRI. In this work, we present a machine learning method for 3D reconstruction and analysis of portable LF-MRI across a range of contrasts and resolutions. Our method works "out of the box" without retraining. It uses a 3D U-Net trained on synthetic LF-MRI to predict signed distance functions of cortical surfaces, followed by geometric processing to ensure topological accuracy. We evaluate our method using paired HF/LF-MRI scans of the same subjects, showing that LF-MRI surface reconstruction accuracy depends on acquisition parameters, including contrast type (T1 vs T2), orientation (axial vs isotropic), and resolution. A 3mm isotropic T2-weighted scan acquired in under 4 minutes, yields strong agreement with HF-derived surfaces: surface area correlates at r=0.96, cortical parcellations reach Dice=0.98, and gray matter volume achieves r=0.93. Cortical thickness remains more challenging with correlations up to r=0.70, reflecting the difficulty of sub-mm precision with 3mm voxels. We further validate our method on challenging postmortem LF-MRI, demonstrating its robustness. Our method represents a step toward enabling cortical surface analysis on portable LF-MRI. Code is available at https://surfer.nmr.mgh.harvard.edu/fswiki/ReconAny
Deep learning of personalized priors from past MRI scans enables fast, quality-enhanced point-of-care MRI with low-cost systems
May 05, 2025Abstract:Magnetic resonance imaging (MRI) offers superb-quality images, but its accessibility is limited by high costs, posing challenges for patients requiring longitudinal care. Low-field MRI provides affordable imaging with low-cost devices but is hindered by long scans and degraded image quality, including low signal-to-noise ratio (SNR) and tissue contrast. We propose a novel healthcare paradigm: using deep learning to extract personalized features from past standard high-field MRI scans and harnessing them to enable accelerated, enhanced-quality follow-up scans with low-cost systems. To overcome the SNR and contrast differences, we introduce ViT-Fuser, a feature-fusion vision transformer that learns features from past scans, e.g. those stored in standard DICOM CDs. We show that \textit{a single prior scan is sufficient}, and this scan can come from various MRI vendors, field strengths, and pulse sequences. Experiments with four datasets, including glioblastoma data, low-field ($50mT$), and ultra-low-field ($6.5mT$) data, demonstrate that ViT-Fuser outperforms state-of-the-art methods, providing enhanced-quality images from accelerated low-field scans, with robustness to out-of-distribution data. Our freely available framework thus enables rapid, diagnostic-quality, low-cost imaging for wide healthcare applications.
Reference-Free 3D Reconstruction of Brain Dissection Photographs with Machine Learning
Mar 13, 2025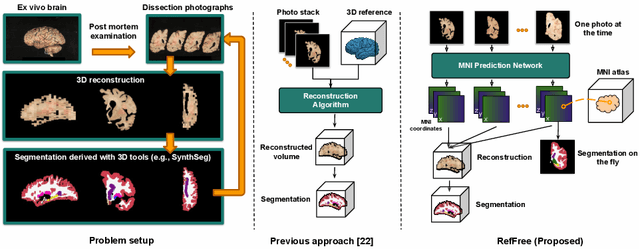
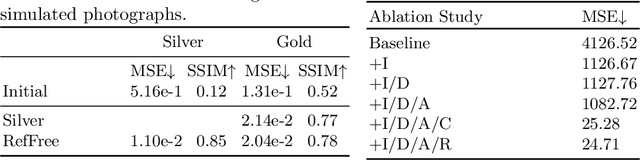
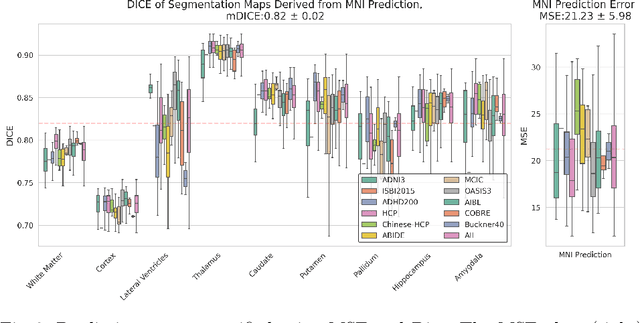
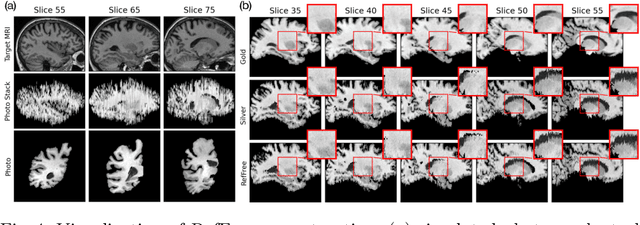
Abstract:Correlation of neuropathology with MRI has the potential to transfer microscopic signatures of pathology to invivo scans. Recently, a classical registration method has been proposed, to build these correlations from 3D reconstructed stacks of dissection photographs, which are routinely taken at brain banks. These photographs bypass the need for exvivo MRI, which is not widely accessible. However, this method requires a full stack of brain slabs and a reference mask (e.g., acquired with a surface scanner), which severely limits the applicability of the technique. Here we propose RefFree, a dissection photograph reconstruction method without external reference. RefFree is a learning approach that estimates the 3D coordinates in the atlas space for every pixel in every photograph; simple least-squares fitting can then be used to compute the 3D reconstruction. As a by-product, RefFree also produces an atlas-based segmentation of the reconstructed stack. RefFree is trained on synthetic photographs generated from digitally sliced 3D MRI data, with randomized appearance for enhanced generalization ability. Experiments on simulated and real data show that RefFree achieves performance comparable to the baseline method without an explicit reference while also enabling reconstruction of partial stacks. Our code is available at https://github.com/lintian-a/reffree.
Probabilistic Contrastive Learning with Explicit Concentration on the Hypersphere
May 26, 2024Abstract:Self-supervised contrastive learning has predominantly adopted deterministic methods, which are not suited for environments characterized by uncertainty and noise. This paper introduces a new perspective on incorporating uncertainty into contrastive learning by embedding representations within a spherical space, inspired by the von Mises-Fisher distribution (vMF). We introduce an unnormalized form of vMF and leverage the concentration parameter, kappa, as a direct, interpretable measure to quantify uncertainty explicitly. This approach not only provides a probabilistic interpretation of the embedding space but also offers a method to calibrate model confidence against varying levels of data corruption and characteristics. Our empirical results demonstrate that the estimated concentration parameter correlates strongly with the degree of unforeseen data corruption encountered at test time, enables failure analysis, and enhances existing out-of-distribution detection methods.
Quantifying white matter hyperintensity and brain volumes in heterogeneous clinical and low-field portable MRI
Dec 08, 2023
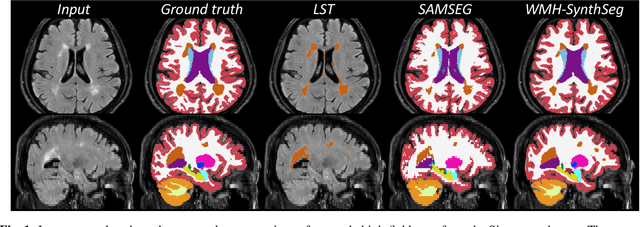

Abstract:Brain atrophy and white matter hyperintensity (WMH) are critical neuroimaging features for ascertaining brain injury in cerebrovascular disease and multiple sclerosis. Automated segmentation and quantification is desirable but existing methods require high-resolution MRI with good signal-to-noise ratio (SNR). This precludes application to clinical and low-field portable MRI (pMRI) scans, thus hampering large-scale tracking of atrophy and WMH progression, especially in underserved areas where pMRI has huge potential. Here we present a method that segments white matter hyperintensity and 36 brain regions from scans of any resolution and contrast (including pMRI) without retraining. We show results on six public datasets and on a private dataset with paired high- and low-field scans (3T and 64mT), where we attain strong correlation between the WMH ($\rho$=.85) and hippocampal volumes (r=.89) estimated at both fields. Our method is publicly available as part of FreeSurfer, at: http://surfer.nmr.mgh.harvard.edu/fswiki/WMH-SynthSeg.
Resolution- and Stimulus-agnostic Super-Resolution of Ultra-High-Field Functional MRI: Application to Visual Studies
Nov 25, 2023



Abstract:High-resolution fMRI provides a window into the brain's mesoscale organization. Yet, higher spatial resolution increases scan times, to compensate for the low signal and contrast-to-noise ratio. This work introduces a deep learning-based 3D super-resolution (SR) method for fMRI. By incorporating a resolution-agnostic image augmentation framework, our method adapts to varying voxel sizes without retraining. We apply this innovative technique to localize fine-scale motion-selective sites in the early visual areas. Detection of these sites typically requires a resolution higher than 1 mm isotropic, whereas here, we visualize them based on lower resolution (2-3mm isotropic) fMRI data. Remarkably, the super-resolved fMRI is able to recover high-frequency detail of the interdigitated organization of these sites (relative to the color-selective sites), even with training data sourced from different subjects and experimental paradigms -- including non-visual resting-state fMRI, underscoring its robustness and versatility. Quantitative and qualitative results indicate that our method has the potential to enhance the spatial resolution of fMRI, leading to a drastic reduction in acquisition time.
Uncertainty Estimation for Deep Learning Image Reconstruction using a Local Lipschitz Metric
May 12, 2023Abstract:The use of deep learning approaches for image reconstruction is of contemporary interest in radiology, especially for approaches that solve inverse problems associated with imaging. In deployment, these models may be exposed to input distributions that are widely shifted from training data, due in part to data biases or drifts. We propose a metric based on local Lipschitz determined from a single trained model that can be used to estimate the model uncertainty for image reconstructions. We demonstrate a monotonic relationship between the local Lipschitz value and Mean Absolute Error and show that this method can be used to provide a threshold that determines whether a given DL reconstruction approach was well suited to the task. Our uncertainty estimation method can be used to identify out-of-distribution test samples, relate information regarding epistemic uncertainties, and guide proper data augmentation. Quantifying uncertainty of learned reconstruction approaches is especially pertinent to the medical domain where reconstructed images must remain diagnostically accurate.
Synthetic Low-Field MRI Super-Resolution Via Nested U-Net Architecture
Nov 28, 2022Abstract:Low-field (LF) MRI scanners have the power to revolutionize medical imaging by providing a portable and cheaper alternative to high-field MRI scanners. However, such scanners are usually significantly noisier and lower quality than their high-field counterparts. The aim of this paper is to improve the SNR and overall image quality of low-field MRI scans to improve diagnostic capability. To address this issue, we propose a Nested U-Net neural network architecture super-resolution algorithm that outperforms previously suggested deep learning methods with an average PSNR of 78.83 and SSIM of 0.9551. We tested our network on artificial noisy downsampled synthetic data from a major T1 weighted MRI image dataset called the T1-mix dataset. One board-certified radiologist scored 25 images on the Likert scale (1-5) assessing overall image quality, anatomical structure, and diagnostic confidence across our architecture and other published works (SR DenseNet, Generator Block, SRCNN, etc.). We also introduce a new type of loss function called natural log mean squared error (NLMSE). In conclusion, we present a more accurate deep learning method for single image super-resolution applied to synthetic low-field MRI via a Nested U-Net architecture.
On Real-time Image Reconstruction with Neural Networks for MRI-guided Radiotherapy
Feb 10, 2022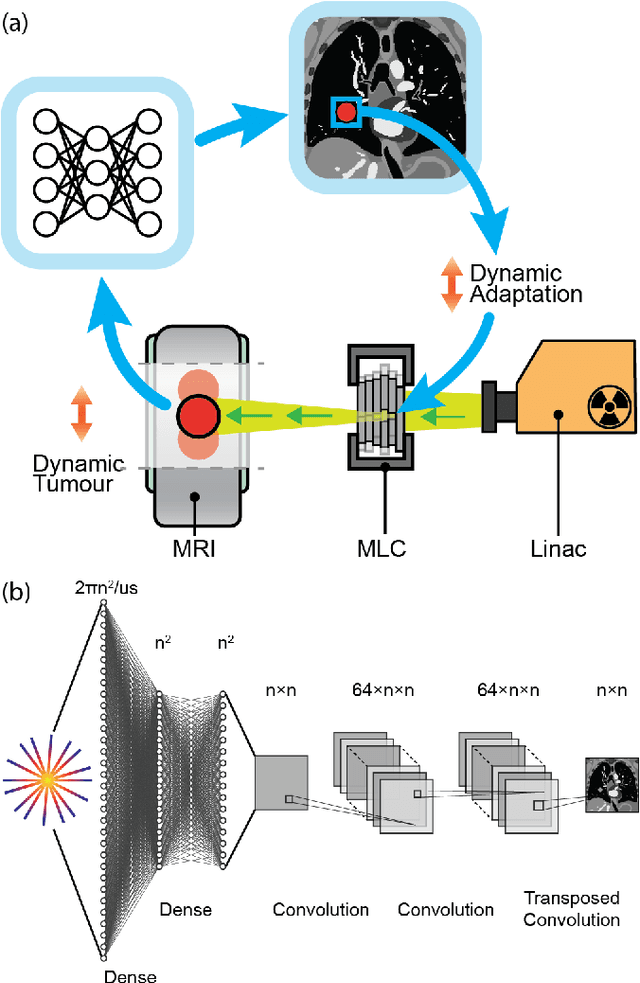
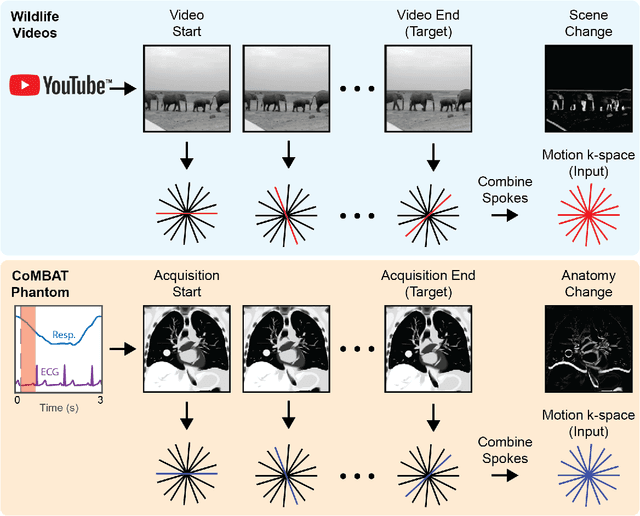
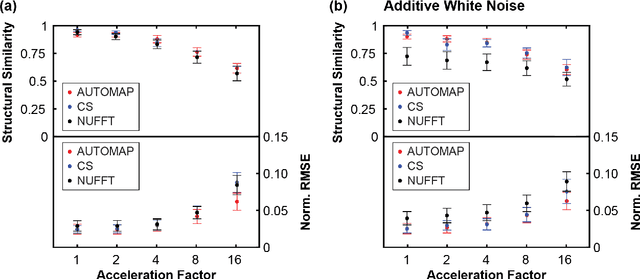
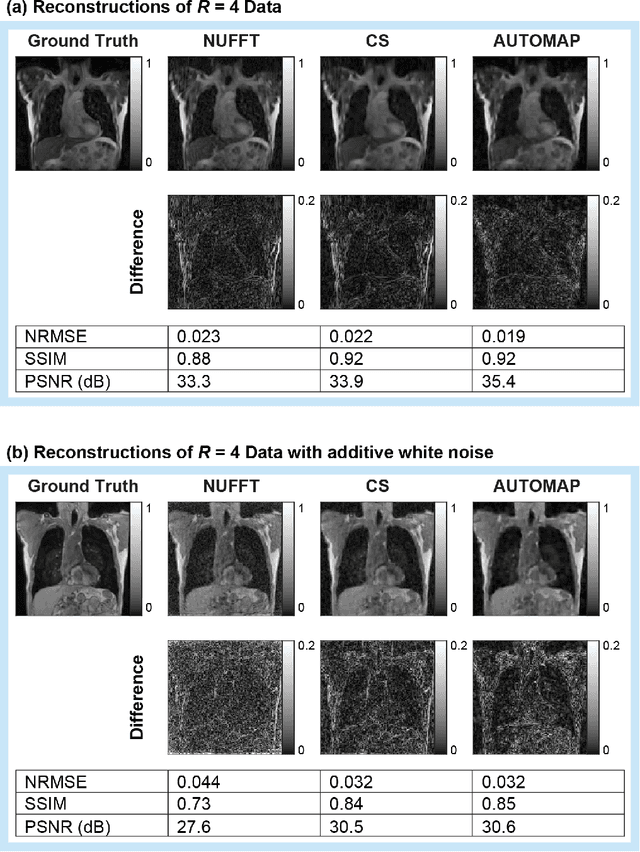
Abstract:MRI-guidance techniques that dynamically adapt radiation beams to follow tumor motion in real-time will lead to more accurate cancer treatments and reduced collateral healthy tissue damage. The gold-standard for reconstruction of undersampled MR data is compressed sensing (CS) which is computationally slow and limits the rate that images can be available for real-time adaptation. Here, we demonstrate the use of automated transform by manifold approximation (AUTOMAP), a generalized framework that maps raw MR signal to the target image domain, to rapidly reconstruct images from undersampled radial k-space data. The AUTOMAP neural network was trained to reconstruct images from a golden-angle radial acquisition, a benchmark for motion-sensitive imaging, on lung cancer patient data and generic images from ImageNet. Model training was subsequently augmented with motion-encoded k-space data derived from videos in the YouTube-8M dataset to encourage motion robust reconstruction. We find that AUTOMAP-reconstructed radial k-space has equivalent accuracy to CS but with much shorter processing times after initial fine-tuning on retrospectively acquired lung cancer patient data. Validation of motion-trained models with a virtual dynamic lung tumor phantom showed that the generalized motion properties learned from YouTube lead to improved target tracking accuracy. Our work shows that AUTOMAP can achieve real-time, accurate reconstruction of radial data. These findings imply that neural-network-based reconstruction is potentially superior to existing approaches for real-time image guidance applications.
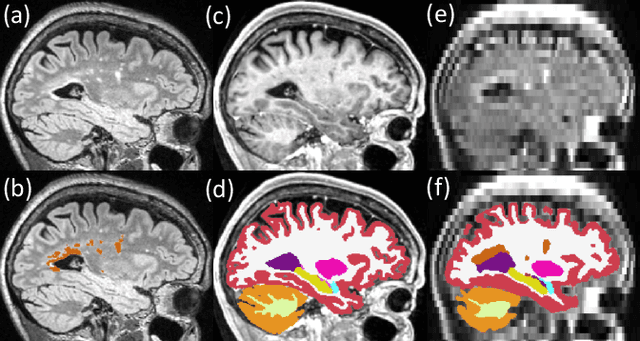
Accurate super-resolution low-field brain MRI
Feb 07, 2022Abstract:The recent introduction of portable, low-field MRI (LF-MRI) into the clinical setting has the potential to transform neuroimaging. However, LF-MRI is limited by lower resolution and signal-to-noise ratio, leading to incomplete characterization of brain regions. To address this challenge, recent advances in machine learning facilitate the synthesis of higher resolution images derived from one or multiple lower resolution scans. Here, we report the extension of a machine learning super-resolution (SR) algorithm to synthesize 1 mm isotropic MPRAGE-like scans from LF-MRI T1-weighted and T2-weighted sequences. Our initial results on a paired dataset of LF and high-field (HF, 1.5T-3T) clinical scans show that: (i) application of available automated segmentation tools directly to LF-MRI images falters; but (ii) segmentation tools succeed when applied to SR images with high correlation to gold standard measurements from HF-MRI (e.g., r = 0.85 for hippocampal volume, r = 0.84 for the thalamus, r = 0.92 for the whole cerebrum). This work demonstrates proof-of-principle post-processing image enhancement from lower resolution LF-MRI sequences. These results lay the foundation for future work to enhance the detection of normal and abnormal image findings at LF and ultimately improve the diagnostic performance of LF-MRI. Our tools are publicly available on FreeSurfer (surfer.nmr.mgh.harvard.edu/).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge