Murthy Devarakonda
sc-OTGM: Single-Cell Perturbation Modeling by Solving Optimal Mass Transport on the Manifold of Gaussian Mixtures
May 06, 2024



Abstract:Influenced by breakthroughs in LLMs, single-cell foundation models are emerging. While these models show successful performance in cell type clustering, phenotype classification, and gene perturbation response prediction, it remains to be seen if a simpler model could achieve comparable or better results, especially with limited data. This is important, as the quantity and quality of single-cell data typically fall short of the standards in textual data used for training LLMs. Single-cell sequencing often suffers from technical artifacts, dropout events, and batch effects. These challenges are compounded in a weakly supervised setting, where the labels of cell states can be noisy, further complicating the analysis. To tackle these challenges, we present sc-OTGM, streamlined with less than 500K parameters, making it approximately 100x more compact than the foundation models, offering an efficient alternative. sc-OTGM is an unsupervised model grounded in the inductive bias that the scRNAseq data can be generated from a combination of the finite multivariate Gaussian distributions. The core function of sc-OTGM is to create a probabilistic latent space utilizing a GMM as its prior distribution and distinguish between distinct cell populations by learning their respective marginal PDFs. It uses a Hit-and-Run Markov chain sampler to determine the OT plan across these PDFs within the GMM framework. We evaluated our model against a CRISPR-mediated perturbation dataset, called CROP-seq, consisting of 57 one-gene perturbations. Our results demonstrate that sc-OTGM is effective in cell state classification, aids in the analysis of differential gene expression, and ranks genes for target identification through a recommender system. It also predicts the effects of single-gene perturbations on downstream gene regulation and generates synthetic scRNA-seq data conditioned on specific cell states.
Customizing Knowledge Graph Embedding to Improve Clinical Study Recommendation
Dec 28, 2022Abstract:Inferring knowledge from clinical trials using knowledge graph embedding is an emerging area. However, customizing graph embeddings for different use cases remains a significant challenge. We propose custom2vec, an algorithmic framework to customize graph embeddings by incorporating user preferences in training the embeddings. It captures user preferences by adding custom nodes and links derived from manually vetted results of a separate information retrieval method. We propose a joint learning objective to preserve the original network structure while incorporating the user's custom annotations. We hypothesize that the custom training improves user-expected predictions, for example, in link prediction tasks. We demonstrate the effectiveness of custom2vec for clinical trials related to non-small cell lung cancer (NSCLC) with two customization scenarios: recommending immuno-oncology trials evaluating PD-1 inhibitors and exploring similar trials that compare new therapies with a standard of care. The results show that custom2vec training achieves better performance than the conventional training methods. Our approach is a novel way to customize knowledge graph embeddings and enable more accurate recommendations and predictions.
A Scalable AI Approach for Clinical Trial Cohort Optimization
Sep 07, 2021



Abstract:FDA has been promoting enrollment practices that could enhance the diversity of clinical trial populations, through broadening eligibility criteria. However, how to broaden eligibility remains a significant challenge. We propose an AI approach to Cohort Optimization (AICO) through transformer-based natural language processing of the eligibility criteria and evaluation of the criteria using real-world data. The method can extract common eligibility criteria variables from a large set of relevant trials and measure the generalizability of trial designs to real-world patients. It overcomes the scalability limits of existing manual methods and enables rapid simulation of eligibility criteria design for a disease of interest. A case study on breast cancer trial design demonstrates the utility of the method in improving trial generalizability.
COVID-19: Comparative Analysis of Methods for Identifying Articles Related to Therapeutics and Vaccines without Using Labeled Data
Jan 05, 2021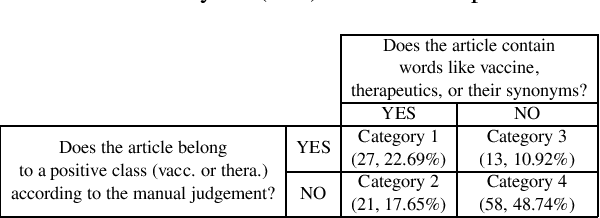
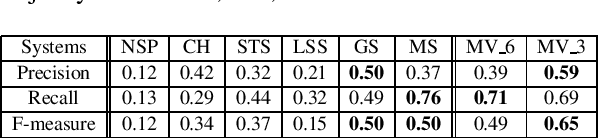
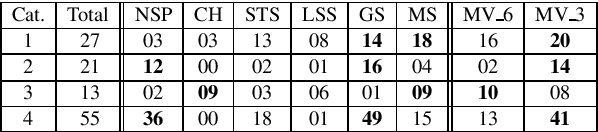
Abstract:Here we proposed an approach to analyze text classification methods based on the presence or absence of task-specific terms (and their synonyms) in the text. We applied this approach to study six different transfer-learning and unsupervised methods for screening articles relevant to COVID-19 vaccines and therapeutics. The analysis revealed that while a BERT model trained on search-engine results generally performed well, it miss-classified relevant abstracts that did not contain task-specific terms. We used this insight to create a more effective unsupervised ensemble.
Cascade Neural Ensemble for Identifying Scientifically Sound Articles
Apr 13, 2020



Abstract:Background: A significant barrier to conducting systematic reviews and meta-analysis is efficiently finding scientifically sound relevant articles. Typically, less than 1% of articles match this requirement which leads to a highly imbalanced task. Although feature-engineered and early neural networks models were studied for this task, there is an opportunity to improve the results. Methods: We framed the problem of filtering articles as a classification task, and trained and tested several ensemble architectures of SciBERT, a variant of BERT pre-trained on scientific articles, on a manually annotated dataset of about 50K articles from MEDLINE. Since scientifically sound articles are identified through a multi-step process we proposed a novel cascade ensemble analogous to the selection process. We compared the performance of the cascade ensemble with a single integrated model and other types of ensembles as well as with results from previous studies. Results: The cascade ensemble architecture achieved 0.7505 F measure, an impressive 49.1% error rate reduction, compared to a CNN model that was previously proposed and evaluated on a selected subset of the 50K articles. On the full dataset, the cascade ensemble achieved 0.7639 F measure, resulting in an error rate reduction of 19.7% compared to the best performance reported in a previous study that used the full dataset. Conclusion: Pre-trained contextual encoder neural networks (e.g. SciBERT) perform better than the models studied previously and manually created search filters in filtering for scientifically sound relevant articles. The superior performance achieved by the cascade ensemble is a significant result that generalizes beyond this task and the dataset, and is analogous to query optimization in IR and databases.
Robustly Pre-trained Neural Model for Direct Temporal Relation Extraction
Apr 13, 2020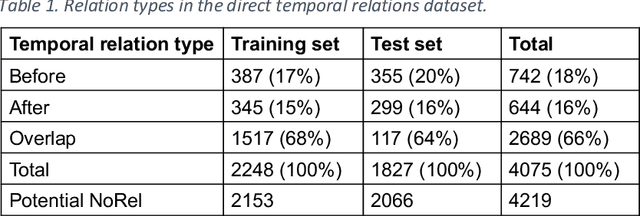
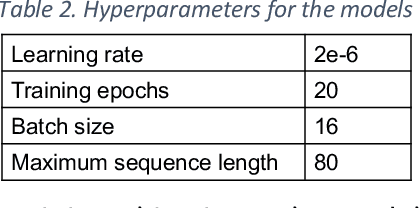
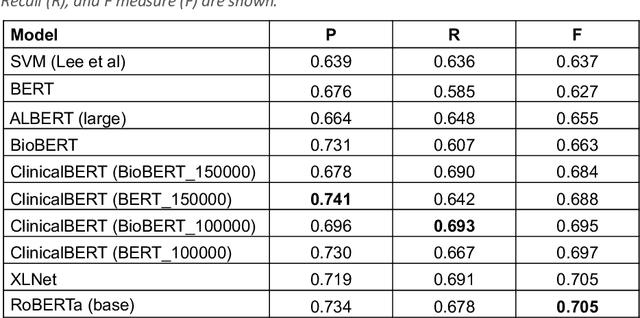
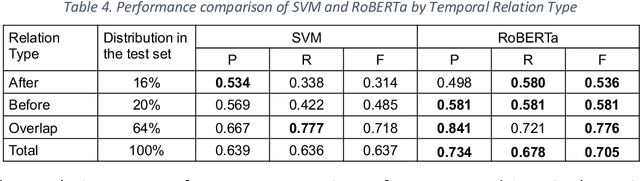
Abstract:Background: Identifying relationships between clinical events and temporal expressions is a key challenge in meaningfully analyzing clinical text for use in advanced AI applications. While previous studies exist, the state-of-the-art performance has significant room for improvement. Methods: We studied several variants of BERT (Bidirectional Encoder Representations using Transformers) some involving clinical domain customization and the others involving improved architecture and/or training strategies. We evaluated these methods using a direct temporal relations dataset which is a semantically focused subset of the 2012 i2b2 temporal relations challenge dataset. Results: Our results show that RoBERTa, which employs better pre-training strategies including using 10x larger corpus, has improved overall F measure by 0.0864 absolute score (on the 1.00 scale) and thus reducing the error rate by 24% relative to the previous state-of-the-art performance achieved with an SVM (support vector machine) model. Conclusion: Modern contextual language modeling neural networks, pre-trained on a large corpus, achieve impressive performance even on highly-nuanced clinical temporal relation tasks.
Knowledge Guided Named Entity Recognition
Nov 10, 2019



Abstract:In this work, we try to perform Named Entity Recognition (NER) with external knowledge. We formulate the NER task as a multi-answer question answering (MAQA) task and provide different knowledge contexts, such as entity types, questions, definitions, and definitions with examples. Moreover, the formulation of the task as a MAQA task helps to reduce other errors. This formulation (a) enables systems to jointly learn from varied NER datasets, enabling systems to learn more NER specific features, (b) can use knowledge-text attention to identify words having higher similarity to 'entity type' mentioned in the knowledge, improving performance, (c) reduces confusion in systems by reducing the classes to be predicted, limited to only three (B, I, O), (d) Makes detection of Nested Entities easier. We perform extensive experiments of this Knowledge Guided NER (KGNER) formulation on 15 Biomedical NER datasets, and through these experiments, we see external knowledge helps. We will release the code for dataset conversion and our trained models for replicating experiments.
Training Models to Extract Treatment Plans from Clinical Notes Using Contents of Sections with Headings
Jun 27, 2019



Abstract:Objective: Using natural language processing (NLP) to find sentences that state treatment plans in a clinical note, would automate plan extraction and would further enable their use in tools that help providers and care managers. However, as in the most NLP tasks on clinical text, creating gold standard to train and test NLP models is tedious and expensive. Fortuitously, sometimes but not always clinical notes contain sections with a heading that identifies the section as a plan. Leveraging contents of such labeled sections as a noisy training data, we assessed accuracy of NLP models trained with the data. Methods: We used common variations of plan headings and rule-based heuristics to find plan sections with headings in clinical notes, and we extracted sentences from them and formed a noisy training data of plan sentences. We trained Support Vector Machine (SVM) and Convolutional Neural Network (CNN) models with the data. We measured accuracy of the trained models on the noisy dataset using ten-fold cross validation and separately on a set-aside manually annotated dataset. Results: About 13% of 117,730 clinical notes contained treatment plans sections with recognizable headings in the 1001 longitudinal patient records that were obtained from Cleveland Clinic under an IRB approval. We were able to extract and create a noisy training data of 13,492 plan sentences from the clinical notes. CNN achieved best F measures, 0.91 and 0.97 in the cross-validation and set-aside evaluation experiments respectively. SVM slightly underperformed with F measures of 0.89 and 0.96 in the same experiments. Conclusion: Our study showed that the training supervised learning models using noisy plan sentences was effective in identifying them in all clinical notes. More broadly, sections with informal headings in clinical notes can be a good source for generating effective training data.
Developing and Using Special-Purpose Lexicons for Cohort Selection from Clinical Notes
Feb 26, 2019
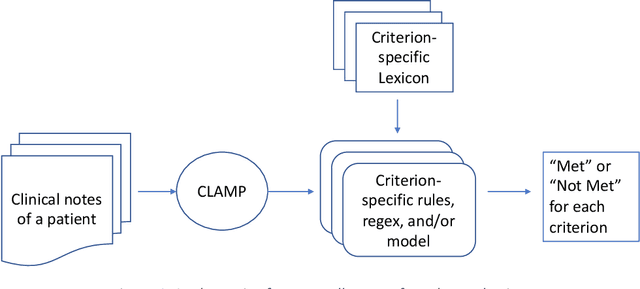

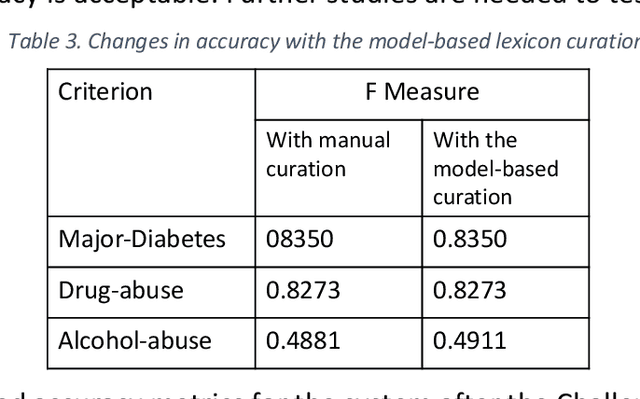
Abstract:Background and Significance: Selecting cohorts for a clinical trial typically requires costly and time-consuming manual chart reviews resulting in poor participation. To help automate the process, National NLP Clinical Challenges (N2C2) conducted a shared challenge by defining 13 criteria for clinical trial cohort selection and by providing training and test datasets. This research was motivated by the N2C2 challenge. Methods: We broke down the task into 13 independent subtasks corresponding to each criterion and implemented subtasks using rules or a supervised machine learning model. Each task critically depended on knowledge resources in the form of task-specific lexicons, for which we developed a novel model-driven approach. The approach allowed us to first expand the lexicon from a seed set and then remove noise from the list, thus improving the accuracy. Results: Our system achieved an overall F measure of 0.9003 at the challenge, and was statistically tied for the first place out of 45 participants. The model-driven lexicon development and further debugging the rules/code on the training set improved overall F measure to 0.9140, overtaking the best numerical result at the challenge. Discussion: Cohort selection, like phenotype extraction and classification, is amenable to rule-based or simple machine learning methods, however, the lexicons involved, such as medication names or medical terms referring to a medical problem, critically determine the overall accuracy. Automated lexicon development has the potential for scalability and accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge