Iya Khalil
TEDDY: A Family Of Foundation Models For Understanding Single Cell Biology
Mar 05, 2025Abstract:Understanding the biological mechanism of disease is critical for medicine, and in particular drug discovery. AI-powered analysis of genome-scale biological data hold great potential in this regard. The increasing availability of single-cell RNA sequencing data has enabled the development of large foundation models for disease biology. However, existing foundation models either do not improve or only modestly improve over task-specific models in downstream applications. Here, we explored two avenues for improving the state-of-the-art. First, we scaled the pre-training dataset to 116 million cells, which is larger than those used by previous models. Second, we leveraged the availability of large-scale biological annotations as a form of supervision during pre-training. We trained the TEDDY family of models comprising six transformer-based state-of-the-art single-cell foundation models with 70 million, 160 million, and 400 million parameters. We vetted our models on two downstream evaluation tasks -- identifying the underlying disease state of held-out donors not seen during training and distinguishing healthy cells from diseased ones for disease conditions and donors not seen during training. Scaling experiments showed that performance improved predictably with both data volume and parameter count. Our models showed substantial improvement over existing work on the first task and more muted improvements on the second.
Customizing Knowledge Graph Embedding to Improve Clinical Study Recommendation
Dec 28, 2022Abstract:Inferring knowledge from clinical trials using knowledge graph embedding is an emerging area. However, customizing graph embeddings for different use cases remains a significant challenge. We propose custom2vec, an algorithmic framework to customize graph embeddings by incorporating user preferences in training the embeddings. It captures user preferences by adding custom nodes and links derived from manually vetted results of a separate information retrieval method. We propose a joint learning objective to preserve the original network structure while incorporating the user's custom annotations. We hypothesize that the custom training improves user-expected predictions, for example, in link prediction tasks. We demonstrate the effectiveness of custom2vec for clinical trials related to non-small cell lung cancer (NSCLC) with two customization scenarios: recommending immuno-oncology trials evaluating PD-1 inhibitors and exploring similar trials that compare new therapies with a standard of care. The results show that custom2vec training achieves better performance than the conventional training methods. Our approach is a novel way to customize knowledge graph embeddings and enable more accurate recommendations and predictions.
EEG-NeXt: A Modernized ConvNet for The Classification of Cognitive Activity from EEG
Dec 08, 2022Abstract:One of the main challenges in electroencephalogram (EEG) based brain-computer interface (BCI) systems is learning the subject/session invariant features to classify cognitive activities within an end-to-end discriminative setting. We propose a novel end-to-end machine learning pipeline, EEG-NeXt, which facilitates transfer learning by: i) aligning the EEG trials from different subjects in the Euclidean-space, ii) tailoring the techniques of deep learning for the scalograms of EEG signals to capture better frequency localization for low-frequency, longer-duration events, and iii) utilizing pretrained ConvNeXt (a modernized ResNet architecture which supersedes state-of-the-art (SOTA) image classification models) as the backbone network via adaptive finetuning. On publicly available datasets (Physionet Sleep Cassette and BNCI2014001) we benchmark our method against SOTA via cross-subject validation and demonstrate improved accuracy in cognitive activity classification along with better generalizability across cohorts.
A Scalable AI Approach for Clinical Trial Cohort Optimization
Sep 07, 2021



Abstract:FDA has been promoting enrollment practices that could enhance the diversity of clinical trial populations, through broadening eligibility criteria. However, how to broaden eligibility remains a significant challenge. We propose an AI approach to Cohort Optimization (AICO) through transformer-based natural language processing of the eligibility criteria and evaluation of the criteria using real-world data. The method can extract common eligibility criteria variables from a large set of relevant trials and measure the generalizability of trial designs to real-world patients. It overcomes the scalability limits of existing manual methods and enables rapid simulation of eligibility criteria design for a disease of interest. A case study on breast cancer trial design demonstrates the utility of the method in improving trial generalizability.
Attention-Based LSTM Network for COVID-19 Clinical Trial Parsing
Dec 18, 2020


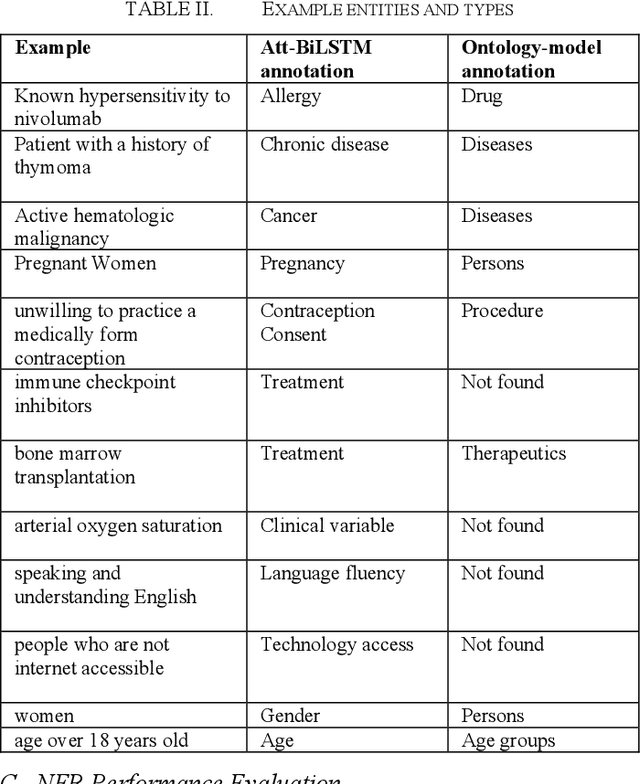
Abstract:COVID-19 clinical trial design is a critical task in developing therapeutics for the prevention and treatment of COVID-19. In this study, we apply a deep learning approach to extract eligibility criteria variables from COVID-19 trials to enable quantitative analysis of trial design and optimization. Specifically, we train attention-based bidirectional Long Short-Term Memory (Att-BiLSTM) models and use the optimal model to extract entities (i.e., variables) from the eligibility criteria of COVID-19 trials. We compare the performance of Att-BiLSTM with traditional ontology-based method. The result on a benchmark dataset shows that Att-BiLSTM outperforms the ontology model. Att-BiLSTM achieves a precision of 0.942, recall of 0.810, and F1 of 0.871, while the ontology model only achieves a precision of 0.715, recall of 0.659, and F1 of 0.686. Our analyses demonstrate that Att-BiLSTM is an effective approach for characterizing patient populations in COVID-19 clinical trials.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge