Xiong Liu
ChARM: Character-based Act-adaptive Reward Modeling for Advanced Role-Playing Language Agents
May 29, 2025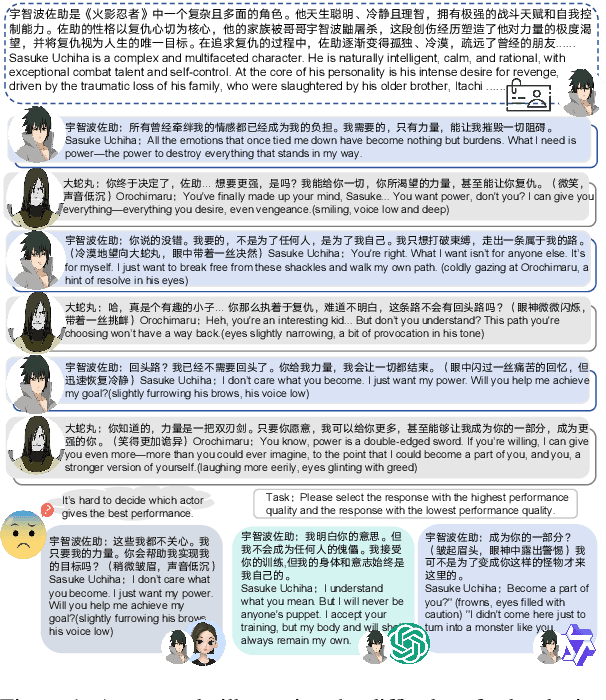
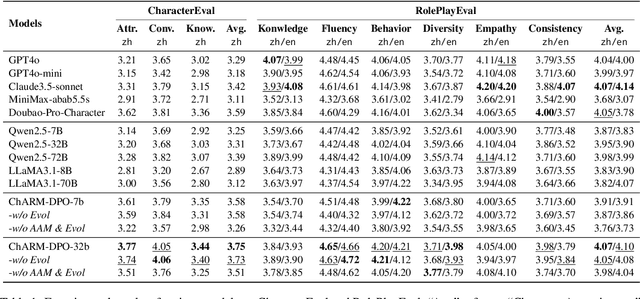
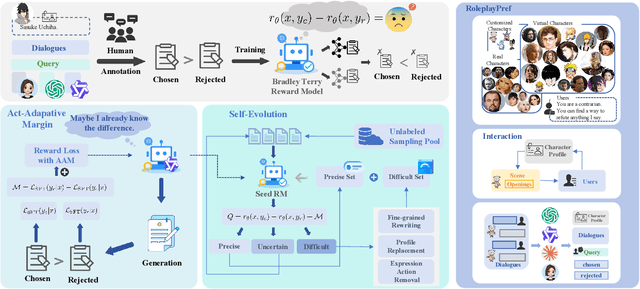
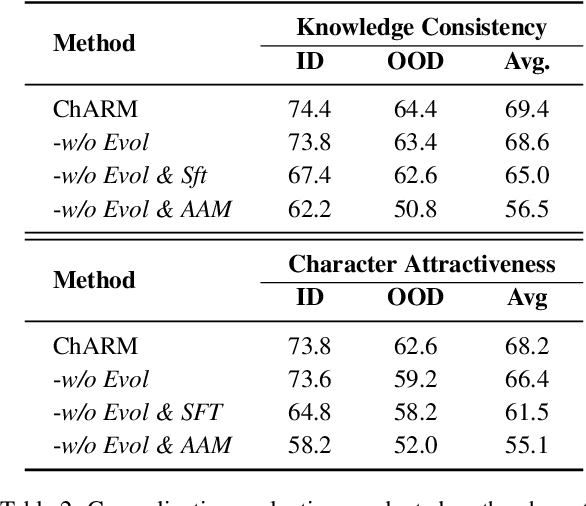
Abstract:Role-Playing Language Agents (RPLAs) aim to simulate characters for realistic and engaging human-computer interactions. However, traditional reward models often struggle with scalability and adapting to subjective conversational preferences. We propose ChARM, a Character-based Act-adaptive Reward Model, addressing these challenges through two innovations: (1) an act-adaptive margin that significantly enhances learning efficiency and generalizability, and (2) a self-evolution mechanism leveraging large-scale unlabeled data to improve training coverage. Additionally, we introduce RoleplayPref, the first large-scale preference dataset specifically for RPLAs, featuring 1,108 characters, 13 subcategories, and 16,888 bilingual dialogues, alongside RoleplayEval, a dedicated evaluation benchmark. Experimental results show a 13% improvement over the conventional Bradley-Terry model in preference rankings. Furthermore, applying ChARM-generated rewards to preference learning techniques (e.g., direct preference optimization) achieves state-of-the-art results on CharacterEval and RoleplayEval. Code and dataset are available at https://github.com/calubkk/ChARM.
OmniCharacter: Towards Immersive Role-Playing Agents with Seamless Speech-Language Personality Interaction
May 26, 2025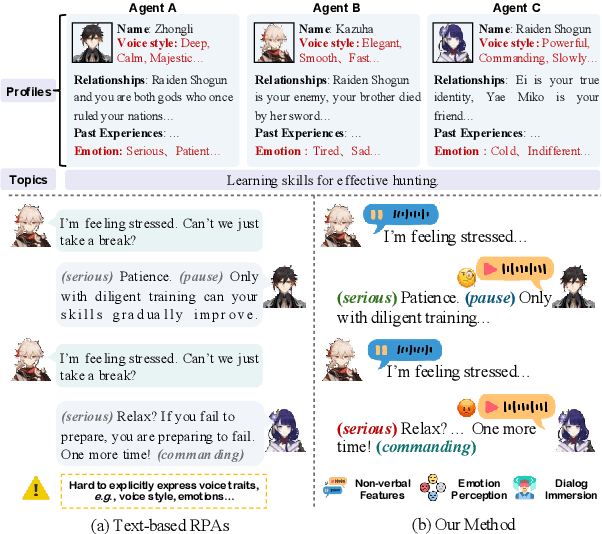



Abstract:Role-Playing Agents (RPAs), benefiting from large language models, is an emerging interactive AI system that simulates roles or characters with diverse personalities. However, existing methods primarily focus on mimicking dialogues among roles in textual form, neglecting the role's voice traits (e.g., voice style and emotions) as playing a crucial effect in interaction, which tends to be more immersive experiences in realistic scenarios. Towards this goal, we propose OmniCharacter, a first seamless speech-language personality interaction model to achieve immersive RPAs with low latency. Specifically, OmniCharacter enables agents to consistently exhibit role-specific personality traits and vocal traits throughout the interaction, enabling a mixture of speech and language responses. To align the model with speech-language scenarios, we construct a dataset named OmniCharacter-10K, which involves more distinctive characters (20), richly contextualized multi-round dialogue (10K), and dynamic speech response (135K). Experimental results showcase that our method yields better responses in terms of both content and style compared to existing RPAs and mainstream speech-language models, with a response latency as low as 289ms. Code and dataset are available at https://github.com/AlibabaResearch/DAMO-ConvAI/tree/main/OmniCharacter.
OpenOmni: Large Language Models Pivot Zero-shot Omnimodal Alignment across Language with Real-time Self-Aware Emotional Speech Synthesis
Jan 08, 2025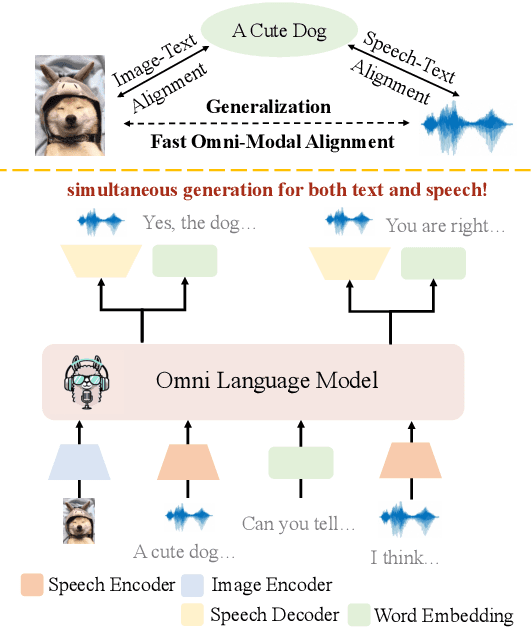
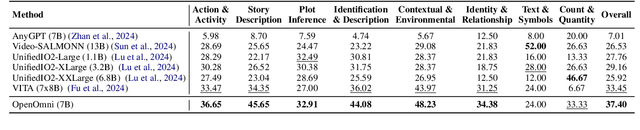
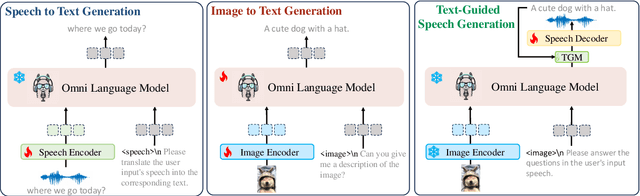
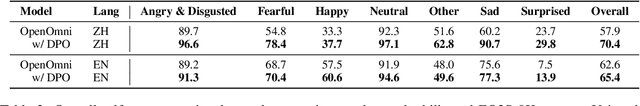
Abstract:Recent advancements in omnimodal learning have been achieved in understanding and generation across images, text, and speech, though mainly within proprietary models. Limited omnimodal datasets and the inherent challenges associated with real-time emotional speech generation have hindered open-source progress. To address these issues, we propose openomni, a two-stage training method combining omnimodal alignment and speech generation to develop a state-of-the-art omnimodal large language model. In the alignment phase, a pre-trained speech model is further trained on text-image tasks to generalize from vision to speech in a (near) zero-shot manner, outperforming models trained on tri-modal datasets. In the speech generation phase, a lightweight decoder facilitates real-time emotional speech through training on speech tasks and preference learning. Experiments demonstrate that openomni consistently improves across omnimodal, vision-language, and speech-language evaluations, enabling natural, emotion-rich dialogues and real-time emotional speech generation.
MMEvol: Empowering Multimodal Large Language Models with Evol-Instruct
Sep 09, 2024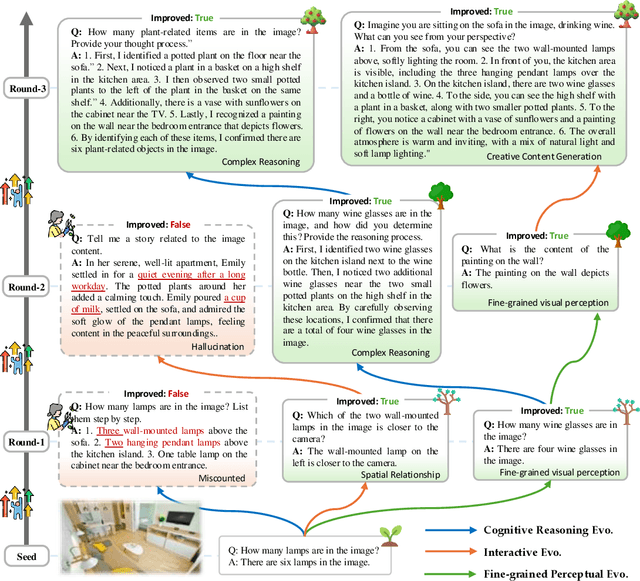
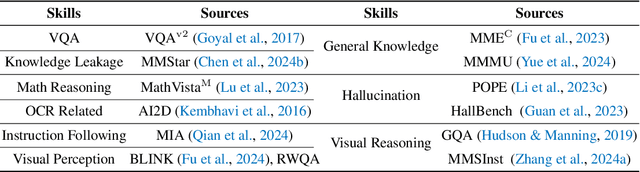
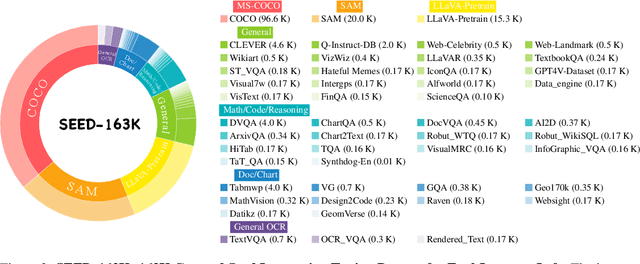
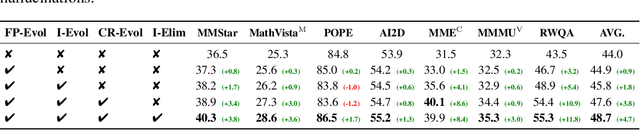
Abstract:The development of Multimodal Large Language Models (MLLMs) has seen significant advancements. However, the quantity and quality of multimodal instruction data have emerged as significant bottlenecks in their progress. Manually creating multimodal instruction data is both time-consuming and inefficient, posing challenges in producing instructions of high complexity. Moreover, distilling instruction data from black-box commercial models (e.g., GPT-4o, GPT-4V) often results in simplistic instruction data, which constrains performance to that of these models. The challenge of curating diverse and complex instruction data remains substantial. We propose MMEvol, a novel multimodal instruction data evolution framework that combines fine-grained perception evolution, cognitive reasoning evolution, and interaction evolution. This iterative approach breaks through data quality bottlenecks to generate a complex and diverse image-text instruction dataset, thereby empowering MLLMs with enhanced capabilities. Beginning with an initial set of instructions, SEED-163K, we utilize MMEvol to systematically broadens the diversity of instruction types, integrates reasoning steps to enhance cognitive capabilities, and extracts detailed information from images to improve visual understanding and robustness. To comprehensively evaluate the effectiveness of our data, we train LLaVA-NeXT using the evolved data and conduct experiments across 13 vision-language tasks. Compared to the baseline trained with seed data, our approach achieves an average accuracy improvement of 3.1 points and reaches state-of-the-art (SOTA) performance on 9 of these tasks.
Improving Factual Consistency of Text Summarization by Adversarially Decoupling Comprehension and Embellishment Abilities of LLMs
Nov 01, 2023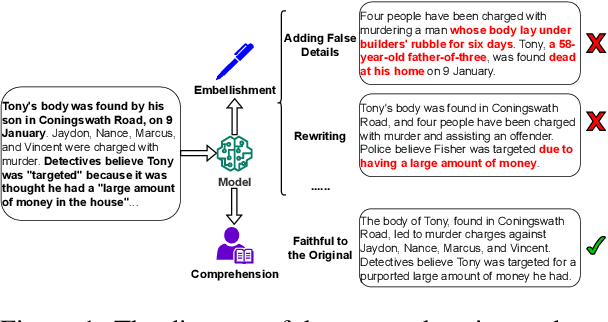
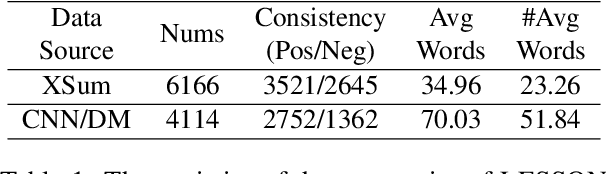
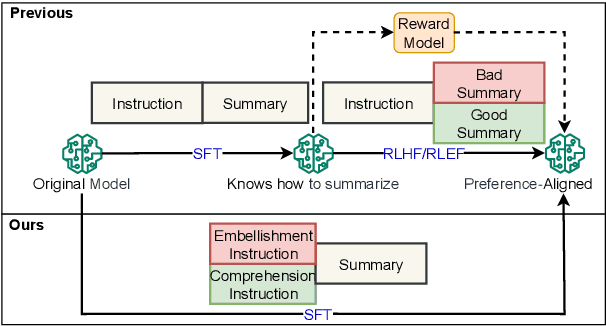
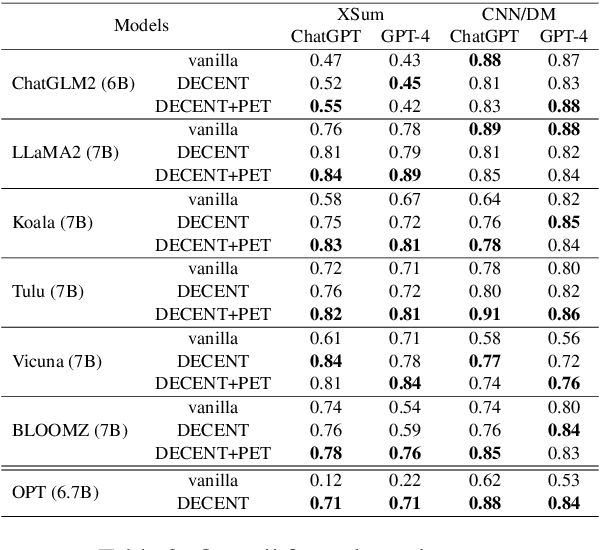
Abstract:Despite the recent progress in text summarization made by large language models (LLMs), they often generate summaries that are factually inconsistent with original articles, known as "hallucinations" in text generation. Unlike previous small models (e.g., BART, T5), current LLMs make fewer silly mistakes but more sophisticated ones, such as imposing cause and effect, adding false details, and overgeneralizing, etc. These hallucinations are challenging to detect through traditional methods, which poses great challenges for improving the factual consistency of text summarization. In this paper, we propose an adversarially DEcoupling method to disentangle the Comprehension and EmbellishmeNT abilities of LLMs (DECENT). Furthermore, we adopt a probing-based parameter-efficient technique to cover the shortage of sensitivity for true and false in the training process of LLMs. In this way, LLMs are less confused about embellishing and understanding, thus can execute the instructions more accurately and have enhanced abilities to distinguish hallucinations. Experimental results show that DECENT significantly improves the reliability of text summarization based on LLMs.
Clinical Trial Recommendations Using Semantics-Based Inductive Inference and Knowledge Graph Embeddings
Sep 27, 2023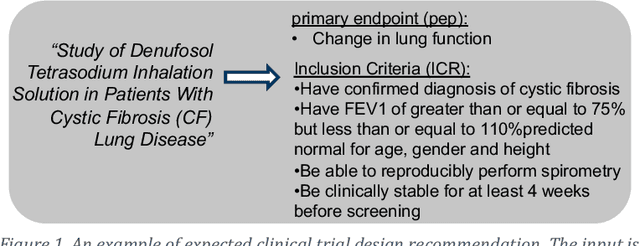
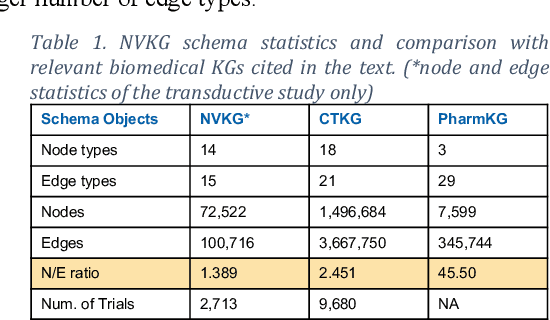
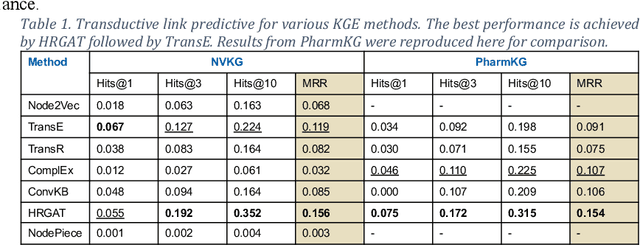
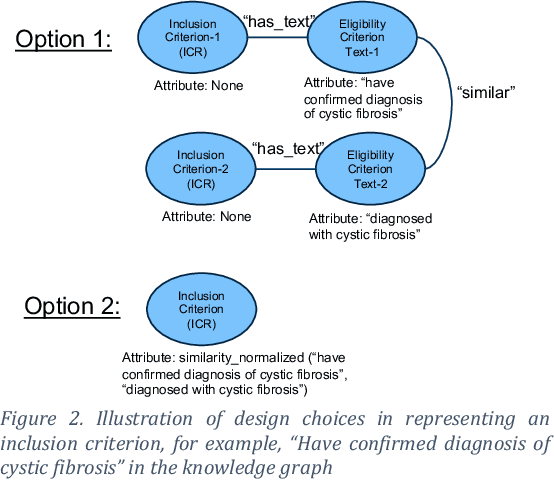
Abstract:Designing a new clinical trial entails many decisions, such as defining a cohort and setting the study objectives to name a few, and therefore can benefit from recommendations based on exhaustive mining of past clinical trial records. Here, we propose a novel recommendation methodology, based on neural embeddings trained on a first-of-a-kind knowledge graph of clinical trials. We addressed several important research questions in this context, including designing a knowledge graph (KG) for clinical trial data, effectiveness of various KG embedding (KGE) methods for it, a novel inductive inference using KGE, and its use in generating recommendations for clinical trial design. We used publicly available data from clinicaltrials.gov for the study. Results show that our recommendations approach achieves relevance scores of 70%-83%, measured as the text similarity to actual clinical trial elements, and the most relevant recommendation can be found near the top of list. Our study also suggests potential improvement in training KGE using node semantics.
Customizing Knowledge Graph Embedding to Improve Clinical Study Recommendation
Dec 28, 2022Abstract:Inferring knowledge from clinical trials using knowledge graph embedding is an emerging area. However, customizing graph embeddings for different use cases remains a significant challenge. We propose custom2vec, an algorithmic framework to customize graph embeddings by incorporating user preferences in training the embeddings. It captures user preferences by adding custom nodes and links derived from manually vetted results of a separate information retrieval method. We propose a joint learning objective to preserve the original network structure while incorporating the user's custom annotations. We hypothesize that the custom training improves user-expected predictions, for example, in link prediction tasks. We demonstrate the effectiveness of custom2vec for clinical trials related to non-small cell lung cancer (NSCLC) with two customization scenarios: recommending immuno-oncology trials evaluating PD-1 inhibitors and exploring similar trials that compare new therapies with a standard of care. The results show that custom2vec training achieves better performance than the conventional training methods. Our approach is a novel way to customize knowledge graph embeddings and enable more accurate recommendations and predictions.
A Scalable AI Approach for Clinical Trial Cohort Optimization
Sep 07, 2021



Abstract:FDA has been promoting enrollment practices that could enhance the diversity of clinical trial populations, through broadening eligibility criteria. However, how to broaden eligibility remains a significant challenge. We propose an AI approach to Cohort Optimization (AICO) through transformer-based natural language processing of the eligibility criteria and evaluation of the criteria using real-world data. The method can extract common eligibility criteria variables from a large set of relevant trials and measure the generalizability of trial designs to real-world patients. It overcomes the scalability limits of existing manual methods and enables rapid simulation of eligibility criteria design for a disease of interest. A case study on breast cancer trial design demonstrates the utility of the method in improving trial generalizability.
Applications of artificial intelligence in drug development using real-world data
Feb 02, 2021



Abstract:The US Food and Drug Administration (FDA) has been actively promoting the use of real-world data (RWD) in drug development. RWD can generate important real-world evidence reflecting the real-world clinical environment where the treatments are used. Meanwhile, artificial intelligence (AI), especially machine- and deep-learning (ML/DL) methods, have been increasingly used across many stages of the drug development process. Advancements in AI have also provided new strategies to analyze large, multidimensional RWD. Thus, we conducted a rapid review of articles from the past 20 years, to provide an overview of the drug development studies that use both AI and RWD. We found that the most popular applications were adverse event detection, trial recruitment, and drug repurposing. Here, we also discuss current research gaps and future opportunities.
The impact of external innovation on new drug approvals: A retrospective analysis
Feb 02, 2021



Abstract:Pharmaceutical companies are relying more often on external sources of innovation to boost their discovery research productivity. However, more in-depth knowledge about how external innovation may translate to successful product launches is still required in order to better understand how to best leverage the innovation ecosystem. We analyzed the pre-approval publication histories for FDA-approved new molecular entities (NMEs) and new biologic entities (NBEs) launched by 13 top research pharma companies during the last decade (2006-2016). We found that academic institutions contributed the majority of pre-approval publications and that publication subject matter is closely aligned with the strengths of the respective innovator. We found this to also be true for candidate drugs terminated in Phase 3, but the volume of literature on these molecules is substantially less than for approved drugs. This may suggest that approved drugs are often associated with a more robust dataset provided by a large number of institutes. Collectively, the results of our analysis support the hypothesis that a collaborative research innovation environment spanning across academia, industry and government is highly conducive to successful drug approvals.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge