Murthy V. Devarakonda
Clinical Trial Recommendations Using Semantics-Based Inductive Inference and Knowledge Graph Embeddings
Sep 27, 2023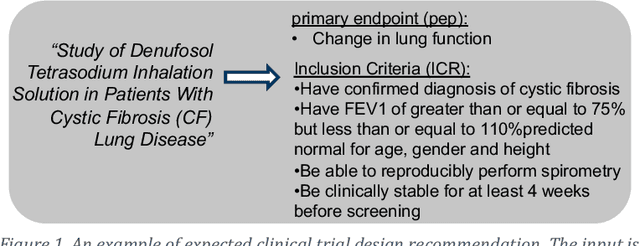
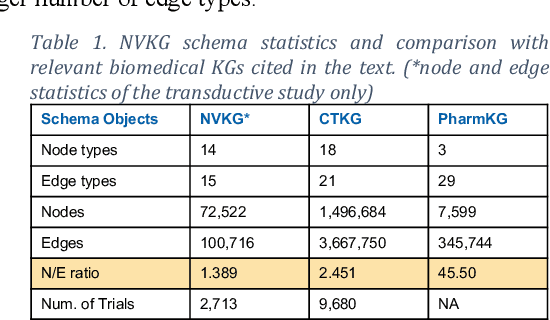
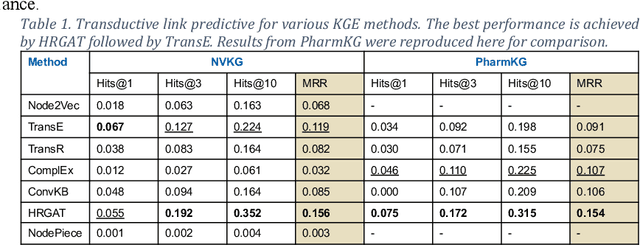
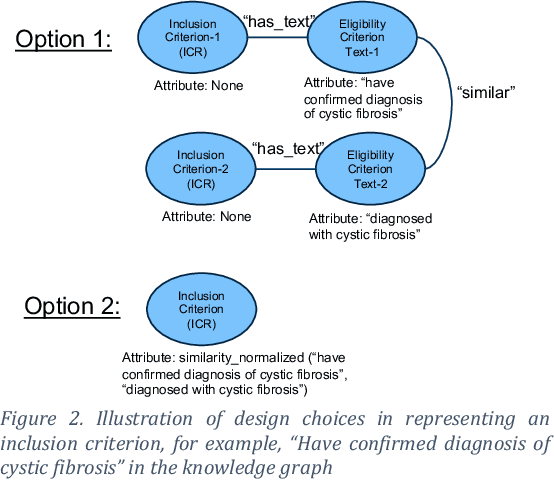
Abstract:Designing a new clinical trial entails many decisions, such as defining a cohort and setting the study objectives to name a few, and therefore can benefit from recommendations based on exhaustive mining of past clinical trial records. Here, we propose a novel recommendation methodology, based on neural embeddings trained on a first-of-a-kind knowledge graph of clinical trials. We addressed several important research questions in this context, including designing a knowledge graph (KG) for clinical trial data, effectiveness of various KG embedding (KGE) methods for it, a novel inductive inference using KGE, and its use in generating recommendations for clinical trial design. We used publicly available data from clinicaltrials.gov for the study. Results show that our recommendations approach achieves relevance scores of 70%-83%, measured as the text similarity to actual clinical trial elements, and the most relevant recommendation can be found near the top of list. Our study also suggests potential improvement in training KGE using node semantics.
Annotating Electronic Medical Records for Question Answering
May 17, 2018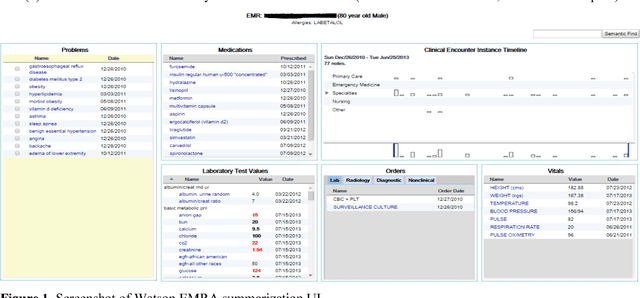
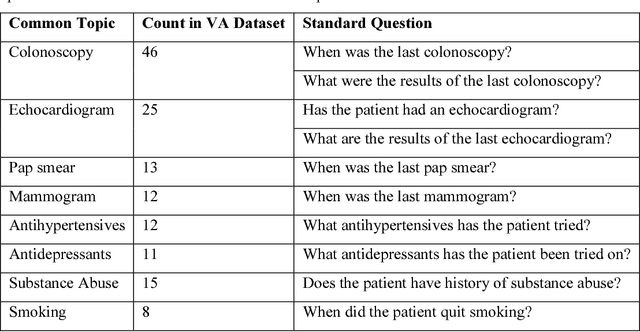
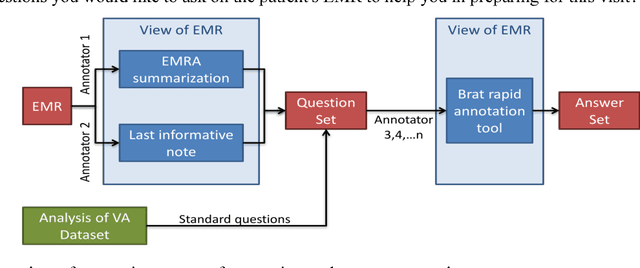
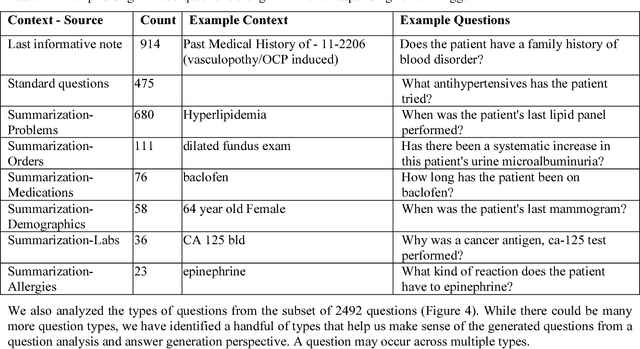
Abstract:Our research is in the relatively unexplored area of question answering technologies for patient-specific questions over their electronic health records. A large dataset of human expert curated question and answer pairs is an important pre-requisite for developing, training and evaluating any question answering system that is powered by machine learning. In this paper, we describe a process for creating such a dataset of questions and answers. Our methodology is replicable, can be conducted by medical students as annotators, and results in high inter-annotator agreement (0.71 Cohen's kappa). Over the course of 11 months, 11 medical students followed our annotation methodology, resulting in a question answering dataset of 5696 questions over 71 patient records, of which 1747 questions have corresponding answers generated by the medical students.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge