William Hogan
Entangled Relations: Leveraging NLI and Meta-analysis to Enhance Biomedical Relation Extraction
May 31, 2024


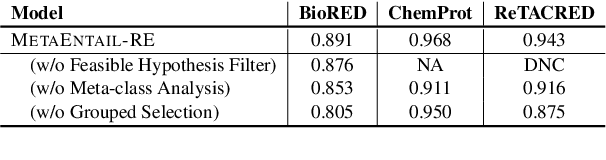
Abstract:Recent research efforts have explored the potential of leveraging natural language inference (NLI) techniques to enhance relation extraction (RE). In this vein, we introduce MetaEntail-RE, a novel adaptation method that harnesses NLI principles to enhance RE performance. Our approach follows past works by verbalizing relation classes into class-indicative hypotheses, aligning a traditionally multi-class classification task to one of textual entailment. We introduce three key enhancements: (1) Instead of labeling non-entailed premise-hypothesis pairs with the uninformative "neutral" entailment label, we introduce meta-class analysis, which provides additional context by analyzing overarching meta relationships between classes when assigning entailment labels; (2) Feasible hypothesis filtering, which removes unlikely hypotheses from consideration based on pairs of entity types; and (3) Group-based prediction selection, which further improves performance by selecting highly confident predictions. MetaEntail-RE is conceptually simple and empirically powerful, yielding significant improvements over conventional relation extraction techniques and other NLI formulations. Our experimental results underscore the versatility of MetaEntail-RE, demonstrating performance gains across both biomedical and general domains.
Precision Rehabilitation for Patients Post-Stroke based on Electronic Health Records and Machine Learning
May 09, 2024



Abstract:In this study, we utilized statistical analysis and machine learning methods to examine whether rehabilitation exercises can improve patients post-stroke functional abilities, as well as forecast the improvement in functional abilities. Our dataset is patients' rehabilitation exercises and demographic information recorded in the unstructured electronic health records (EHRs) data and free-text rehabilitation procedure notes. We collected data for 265 stroke patients from the University of Pittsburgh Medical Center. We employed a pre-existing natural language processing (NLP) algorithm to extract data on rehabilitation exercises and developed a rule-based NLP algorithm to extract Activity Measure for Post-Acute Care (AM-PAC) scores, covering basic mobility (BM) and applied cognitive (AC) domains, from procedure notes. Changes in AM-PAC scores were classified based on the minimal clinically important difference (MCID), and significance was assessed using Friedman and Wilcoxon tests. To identify impactful exercises, we used Chi-square tests, Fisher's exact tests, and logistic regression for odds ratios. Additionally, we developed five machine learning models-logistic regression (LR), Adaboost (ADB), support vector machine (SVM), gradient boosting (GB), and random forest (RF)-to predict outcomes in functional ability. Statistical analyses revealed significant associations between functional improvements and specific exercises. The RF model achieved the best performance in predicting functional outcomes. In this study, we identified three rehabilitation exercises that significantly contributed to patient post-stroke functional ability improvement in the first two months. Additionally, the successful application of a machine learning model to predict patient-specific functional outcomes underscores the potential for precision rehabilitation.
READ: Improving Relation Extraction from an ADversarial Perspective
Apr 02, 2024Abstract:Recent works in relation extraction (RE) have achieved promising benchmark accuracy; however, our adversarial attack experiments show that these works excessively rely on entities, making their generalization capability questionable. To address this issue, we propose an adversarial training method specifically designed for RE. Our approach introduces both sequence- and token-level perturbations to the sample and uses a separate perturbation vocabulary to improve the search for entity and context perturbations. Furthermore, we introduce a probabilistic strategy for leaving clean tokens in the context during adversarial training. This strategy enables a larger attack budget for entities and coaxes the model to leverage relational patterns embedded in the context. Extensive experiments show that compared to various adversarial training methods, our method significantly improves both the accuracy and robustness of the model. Additionally, experiments on different data availability settings highlight the effectiveness of our method in low-resource scenarios. We also perform in-depth analyses of our proposed method and provide further hints. We will release our code at https://github.com/David-Li0406/READ.
Creating a Discipline-specific Commons for Infectious Disease Epidemiology
Nov 12, 2023Abstract:Objective: To create a commons for infectious disease (ID) epidemiology in which epidemiologists, public health officers, data producers, and software developers can not only share data and software, but receive assistance in improving their interoperability. Materials and Methods: We represented 586 datasets, 54 software, and 24 data formats in OWL 2 and then used logical queries to infer potentially interoperable combinations of software and datasets, as well as statistics about the FAIRness of the collection. We represented the objects in DATS 2.2 and a software metadata schema of our own design. We used these representations as the basis for the Content, Search, FAIR-o-meter, and Workflow pages that constitute the MIDAS Digital Commons. Results: Interoperability was limited by lack of standardization of input and output formats of software. When formats existed, they were human-readable specifications (22/24; 92%); only 3 formats (13%) had machine-readable specifications. Nevertheless, logical search of a triple store based on named data formats was able to identify scores of potentially interoperable combinations of software and datasets. Discussion: We improved the findability and availability of a sample of software and datasets and developed metrics for assessing interoperability. The barriers to interoperability included poor documentation of software input/output formats and little attention to standardization of most types of data in this field. Conclusion: Centralizing and formalizing the representation of digital objects within a commons promotes FAIRness, enables its measurement over time and the identification of potentially interoperable combinations of data and software.
DAIL: Data Augmentation for In-Context Learning via Self-Paraphrase
Nov 06, 2023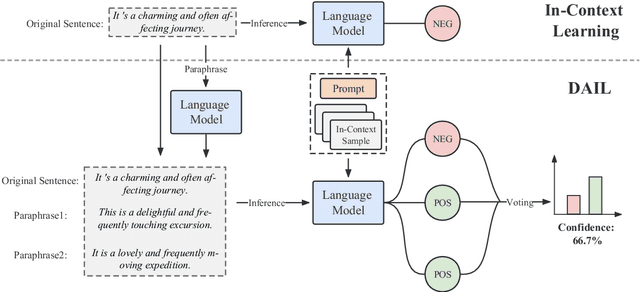
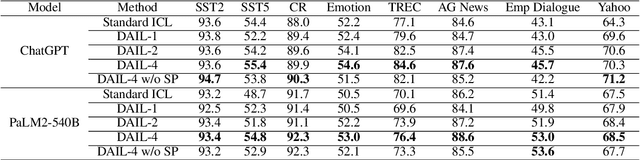
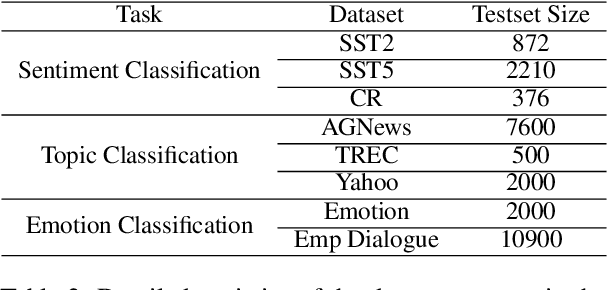
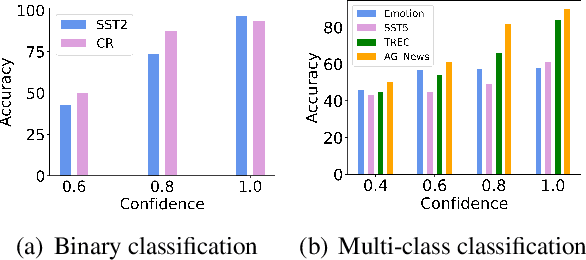
Abstract:In-Context Learning (ICL) combined with pre-trained large language models has achieved promising results on various NLP tasks. However, ICL requires high-quality annotated demonstrations which might not be available in real-world scenarios. To overcome this limitation, we propose \textbf{D}ata \textbf{A}ugmentation for \textbf{I}n-Context \textbf{L}earning (\textbf{DAIL}). DAIL leverages the intuition that large language models are more familiar with the content generated by themselves. It first utilizes the language model to generate paraphrases of the test sample and employs majority voting to determine the final result based on individual predictions. Our extensive empirical evaluation shows that DAIL outperforms the standard ICL method and other ensemble-based methods in the low-resource scenario. Additionally, we explore the use of voting consistency as a confidence score of the model when the logits of predictions are inaccessible. We believe our work will stimulate further research on ICL in low-resource settings.
Open-world Semi-supervised Generalized Relation Discovery Aligned in a Real-world Setting
May 22, 2023Abstract:Open-world Relation Extraction (OpenRE) has recently garnered significant attention. However, existing approaches tend to oversimplify the problem by assuming that all unlabeled texts belong to novel classes, thereby limiting the practicality of these methods. We argue that the OpenRE setting should be more aligned with the characteristics of real-world data. Specifically, we propose two key improvements: (a) unlabeled data should encompass known and novel classes, including hard-negative instances; and (b) the set of novel classes should represent long-tail relation types. Furthermore, we observe that popular relations such as titles and locations can often be implicitly inferred through specific patterns, while long-tail relations tend to be explicitly expressed in sentences. Motivated by these insights, we present a novel method called KNoRD (Known and Novel Relation Discovery), which effectively classifies explicitly and implicitly expressed relations from known and novel classes within unlabeled data. Experimental evaluations on several Open-world RE benchmarks demonstrate that KNoRD consistently outperforms other existing methods, achieving significant performance gains.
An Overview of Distant Supervision for Relation Extraction with a Focus on Denoising and Pre-training Methods
Jul 17, 2022



Abstract:Relation Extraction (RE) is a foundational task of natural language processing. RE seeks to transform raw, unstructured text into structured knowledge by identifying relational information between entity pairs found in text. RE has numerous uses, such as knowledge graph completion, text summarization, question-answering, and search querying. The history of RE methods can be roughly organized into four phases: pattern-based RE, statistical-based RE, neural-based RE, and large language model-based RE. This survey begins with an overview of a few exemplary works in the earlier phases of RE, highlighting limitations and shortcomings to contextualize progress. Next, we review popular benchmarks and critically examine metrics used to assess RE performance. We then discuss distant supervision, a paradigm that has shaped the development of modern RE methods. Lastly, we review recent RE works focusing on denoising and pre-training methods.
Fine-grained Contrastive Learning for Relation Extraction
May 25, 2022



Abstract:Recent relation extraction (RE) works have shown encouraging improvements by conducting contrastive learning on silver labels generated by distant supervision before fine-tuning on gold labels. Existing methods typically assume all these silver labels are accurate and therefore treat them equally in contrastive learning; however, distant supervision is inevitably noisy -- some silver labels are more reliable than others. In this paper, we first assess the quality of silver labels via a simple and automatic approach we call "learning order denoising," where we train a language model to learn these relations and record the order of learned training instances. We show that learning order largely corresponds to label accuracy -- early learned silver labels have, on average, more accurate labels compared to later learned silver labels. We then propose a novel fine-grained contrastive learning (FineCL) for RE, which leverages this additional, fine-grained information about which silver labels are and are not noisy to improve the quality of learned relationship representations for RE. Experiments on many RE benchmarks show consistent, significant performance gains of FineCL over state-of-the-art methods.
Abstractified Multi-instance Learning (AMIL) for Biomedical Relation Extraction
Oct 24, 2021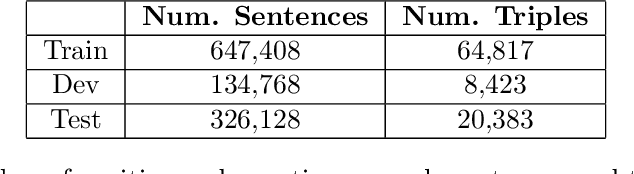

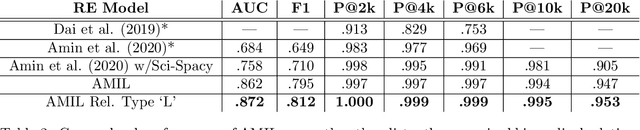
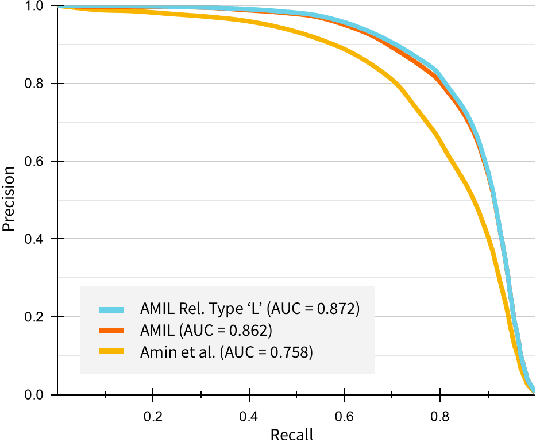
Abstract:Relation extraction in the biomedical domain is a challenging task due to a lack of labeled data and a long-tail distribution of fact triples. Many works leverage distant supervision which automatically generates labeled data by pairing a knowledge graph with raw textual data. Distant supervision produces noisy labels and requires additional techniques, such as multi-instance learning (MIL), to denoise the training signal. However, MIL requires multiple instances of data and struggles with very long-tail datasets such as those found in the biomedical domain. In this work, we propose a novel reformulation of MIL for biomedical relation extraction that abstractifies biomedical entities into their corresponding semantic types. By grouping entities by types, we are better able to take advantage of the benefits of MIL and further denoise the training signal. We show this reformulation, which we refer to as abstractified multi-instance learning (AMIL), improves performance in biomedical relationship extraction. We also propose a novel relationship embedding architecture that further improves model performance.
* 14 pages, 3 figures, submitted to Automated Knowledge Base Construction (2021)
Applications of artificial intelligence in drug development using real-world data
Feb 02, 2021



Abstract:The US Food and Drug Administration (FDA) has been actively promoting the use of real-world data (RWD) in drug development. RWD can generate important real-world evidence reflecting the real-world clinical environment where the treatments are used. Meanwhile, artificial intelligence (AI), especially machine- and deep-learning (ML/DL) methods, have been increasingly used across many stages of the drug development process. Advancements in AI have also provided new strategies to analyze large, multidimensional RWD. Thus, we conducted a rapid review of articles from the past 20 years, to provide an overview of the drug development studies that use both AI and RWD. We found that the most popular applications were adverse event detection, trial recruitment, and drug repurposing. Here, we also discuss current research gaps and future opportunities.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge