Ho-Cheol Kim
Representing Knowledge by Spans: A Knowledge-Enhanced Model for Information Extraction
Aug 20, 2022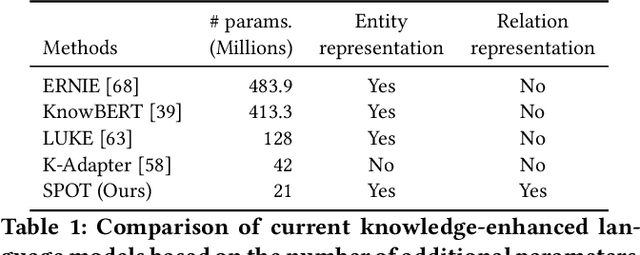
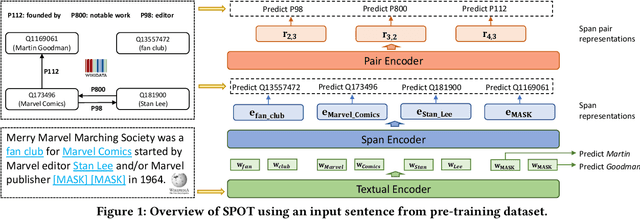
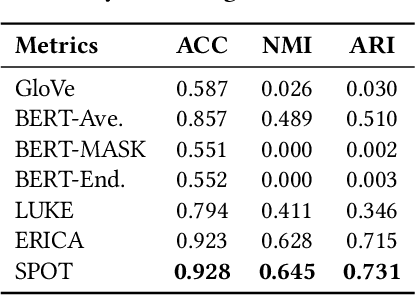
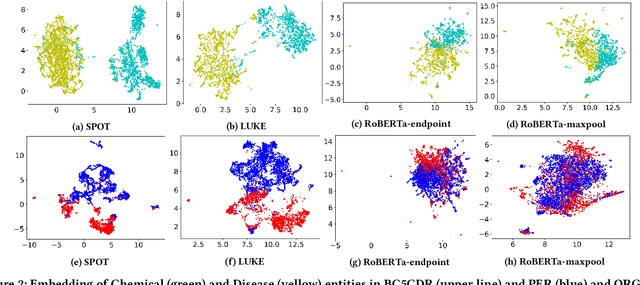
Abstract:Knowledge-enhanced pre-trained models for language representation have been shown to be more effective in knowledge base construction tasks (i.e.,~relation extraction) than language models such as BERT. These knowledge-enhanced language models incorporate knowledge into pre-training to generate representations of entities or relationships. However, existing methods typically represent each entity with a separate embedding. As a result, these methods struggle to represent out-of-vocabulary entities and a large amount of parameters, on top of their underlying token models (i.e.,~the transformer), must be used and the number of entities that can be handled is limited in practice due to memory constraints. Moreover, existing models still struggle to represent entities and relationships simultaneously. To address these problems, we propose a new pre-trained model that learns representations of both entities and relationships from token spans and span pairs in the text respectively. By encoding spans efficiently with span modules, our model can represent both entities and their relationships but requires fewer parameters than existing models. We pre-trained our model with the knowledge graph extracted from Wikipedia and test it on a broad range of supervised and unsupervised information extraction tasks. Results show that our model learns better representations for both entities and relationships than baselines, while in supervised settings, fine-tuning our model outperforms RoBERTa consistently and achieves competitive results on information extraction tasks.
Abstractified Multi-instance Learning (AMIL) for Biomedical Relation Extraction
Oct 24, 2021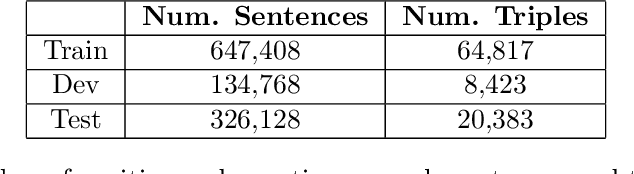

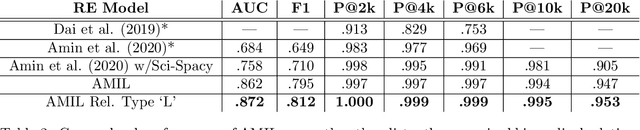
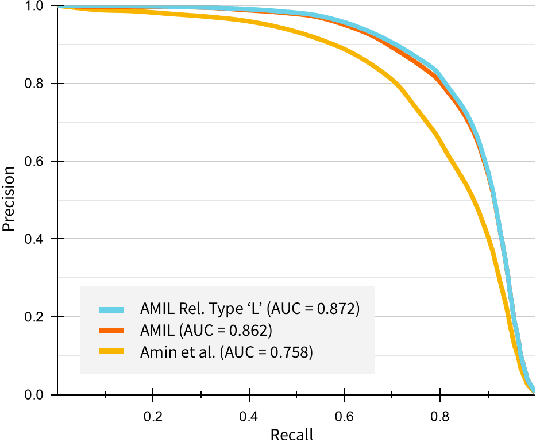
Abstract:Relation extraction in the biomedical domain is a challenging task due to a lack of labeled data and a long-tail distribution of fact triples. Many works leverage distant supervision which automatically generates labeled data by pairing a knowledge graph with raw textual data. Distant supervision produces noisy labels and requires additional techniques, such as multi-instance learning (MIL), to denoise the training signal. However, MIL requires multiple instances of data and struggles with very long-tail datasets such as those found in the biomedical domain. In this work, we propose a novel reformulation of MIL for biomedical relation extraction that abstractifies biomedical entities into their corresponding semantic types. By grouping entities by types, we are better able to take advantage of the benefits of MIL and further denoise the training signal. We show this reformulation, which we refer to as abstractified multi-instance learning (AMIL), improves performance in biomedical relationship extraction. We also propose a novel relationship embedding architecture that further improves model performance.
* 14 pages, 3 figures, submitted to Automated Knowledge Base Construction (2021)
Theoretical Knowledge Graph Reasoning via Ending Anchored Rules
Dec 15, 2020

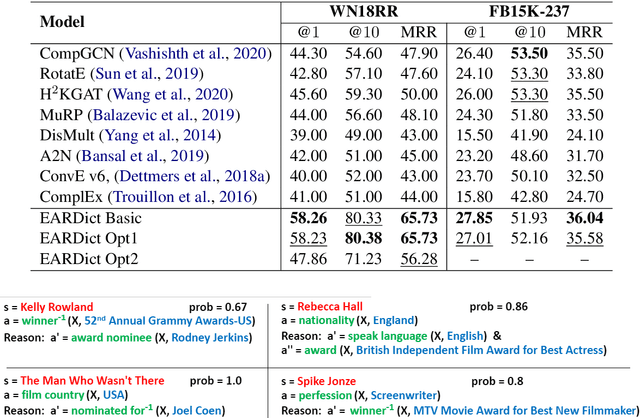
Abstract:Discovering precise and specific rules from knowledge graphs is regarded as an essential challenge, which can improve the performances of many downstream tasks and even provide new ways to approach some Natural Language Processing research topics. In this paper, we provide a fundamental theory for knowledge graph reasoning based on the ending anchored rules. Our theory provides precise reasons explaining why or why not a triple is correct. Then, we implement our theory by what we call the EARDict model. Results show that our EARDict model significantly outperforms all the benchmark models on two large datasets of knowledge graph completion, including achieving a Hits@10 score of 96.6 percent on WN18RR.
Utilizing stability criteria in choosing feature selection methods yields reproducible results in microbiome data
Nov 30, 2020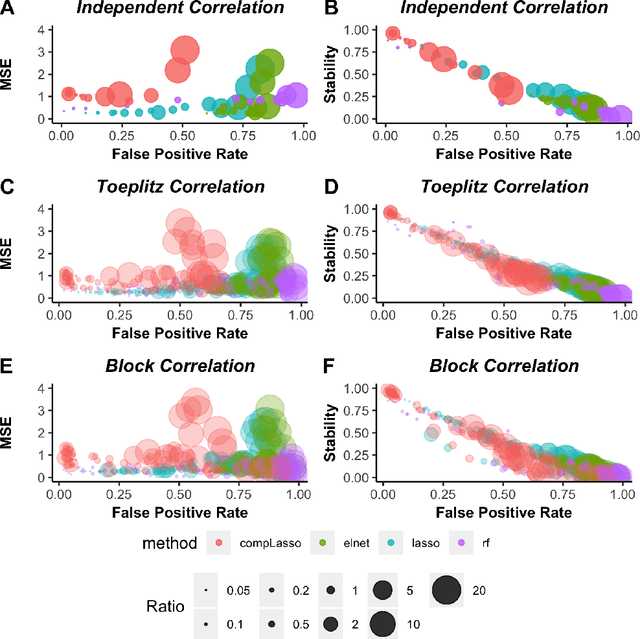
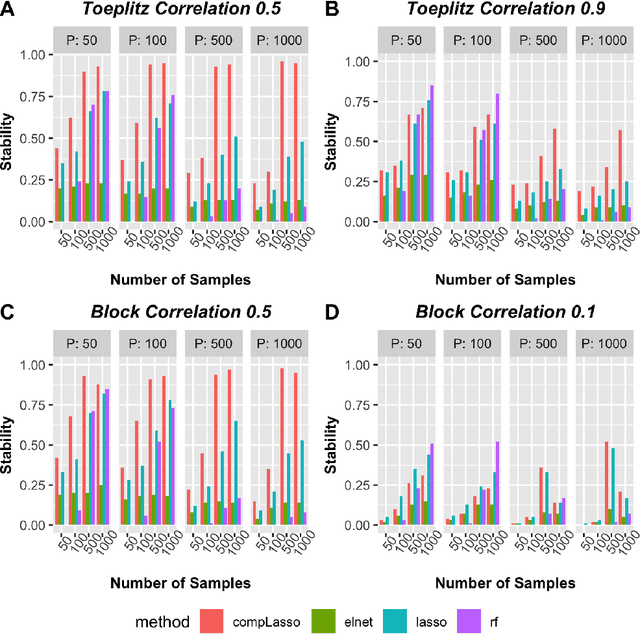
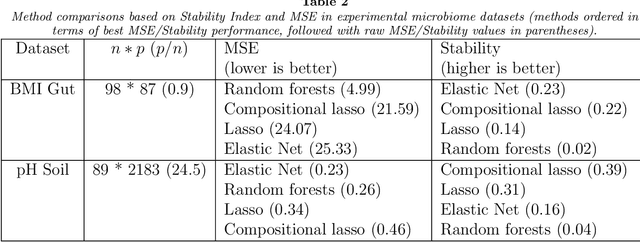
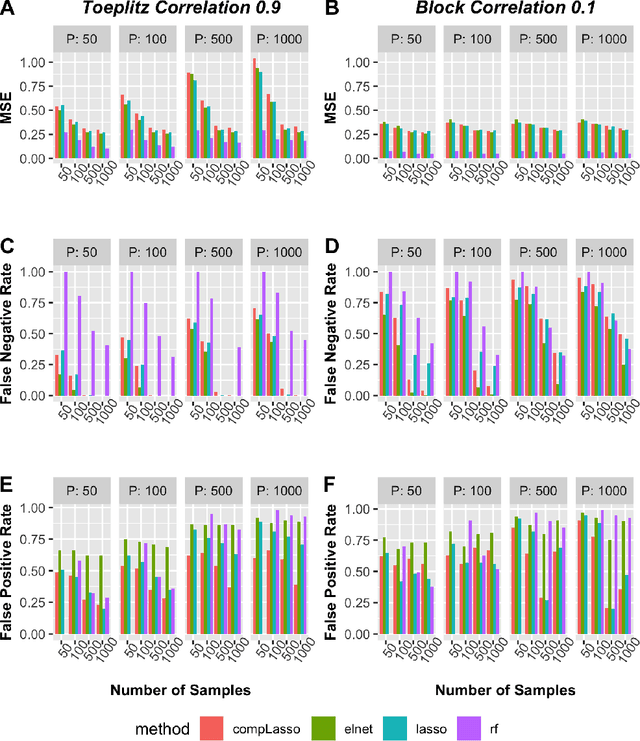
Abstract:Feature selection is indispensable in microbiome data analysis, but it can be particularly challenging as microbiome data sets are high-dimensional, underdetermined, sparse and compositional. Great efforts have recently been made on developing new methods for feature selection that handle the above data characteristics, but almost all methods were evaluated based on performance of model predictions. However, little attention has been paid to address a fundamental question: how appropriate are those evaluation criteria? Most feature selection methods often control the model fit, but the ability to identify meaningful subsets of features cannot be evaluated simply based on the prediction accuracy. If tiny changes to the training data would lead to large changes in the chosen feature subset, then many of the biological features that an algorithm has found are likely to be a data artifact rather than real biological signal. This crucial need of identifying relevant and reproducible features motivated the reproducibility evaluation criterion such as Stability, which quantifies how robust a method is to perturbations in the data. In our paper, we compare the performance of popular model prediction metric MSE and proposed reproducibility criterion Stability in evaluating four widely used feature selection methods in both simulations and experimental microbiome applications. We conclude that Stability is a preferred feature selection criterion over MSE because it better quantifies the reproducibility of the feature selection method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge