Shi Huang
3D Path Planning and Obstacle Avoidance Algorithms for Obstacle-Overcoming Robots
Sep 02, 2022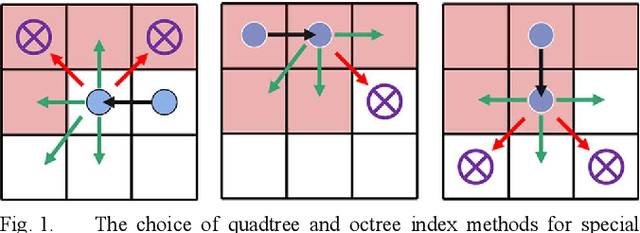
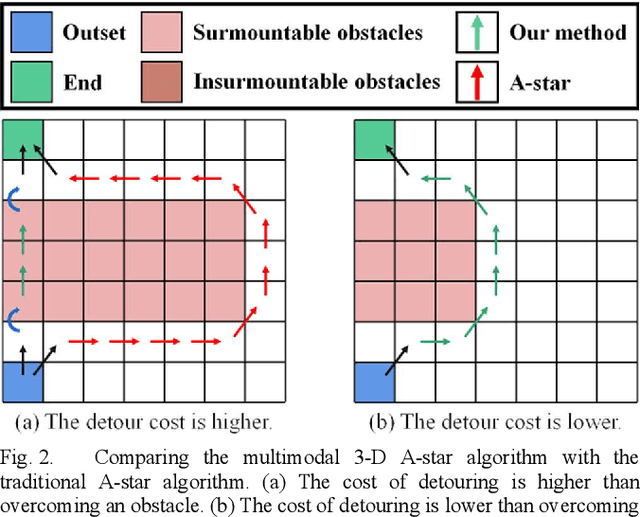
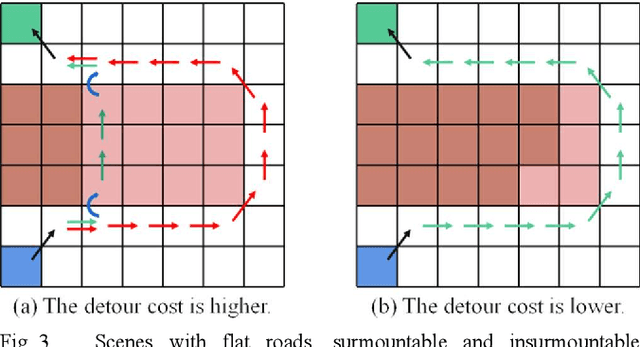
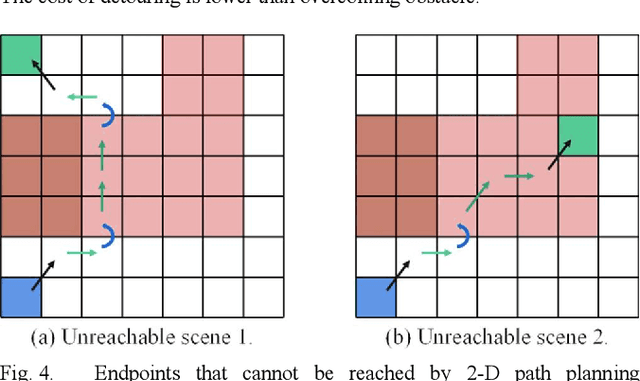
Abstract:This article introduces a multimodal motion planning (MMP) algorithm that combines three-dimensional (3-D) path planning and a DWA obstacle avoidance algorithm. The algorithms aim to plan the path and motion of obstacle-overcoming robots in complex unstructured scenes. A novel A-star algorithm is proposed to combine the characteristics of unstructured scenes and a strategy to switch it into a greedy best-first strategy algorithm. Meanwhile, the algorithm of path planning is integrated with the DWA algorithm so that the robot can perform local dynamic obstacle avoidance during the movement along the global planned path. Furthermore, when the proposed global path planning algorithm combines with the local obstacle avoidance algorithm, the robot can correct the path after obstacle avoidance and obstacle overcoming. The simulation experiments in a factory with several complex environments verified the feasibility and robustness of the algorithms. The algorithms can quickly generate a reasonable 3-D path for obstacle-overcoming robots and perform reliable local obstacle avoidance under the premise of considering the characteristics of the scene and motion obstacles.
Utilizing stability criteria in choosing feature selection methods yields reproducible results in microbiome data
Nov 30, 2020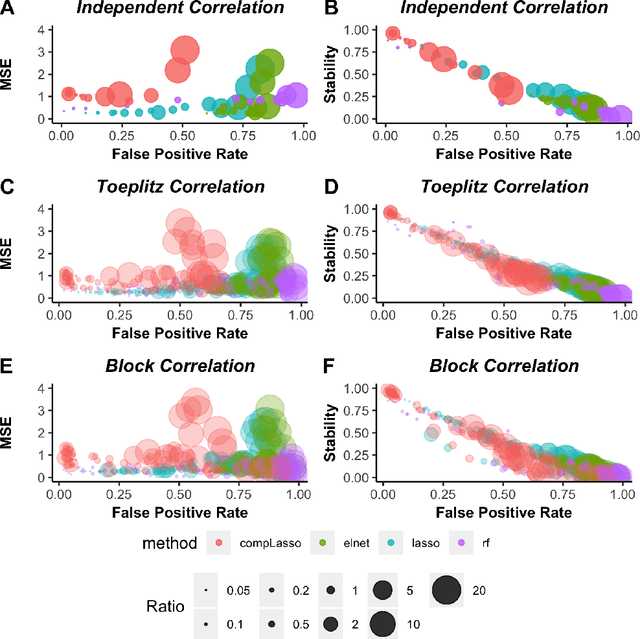
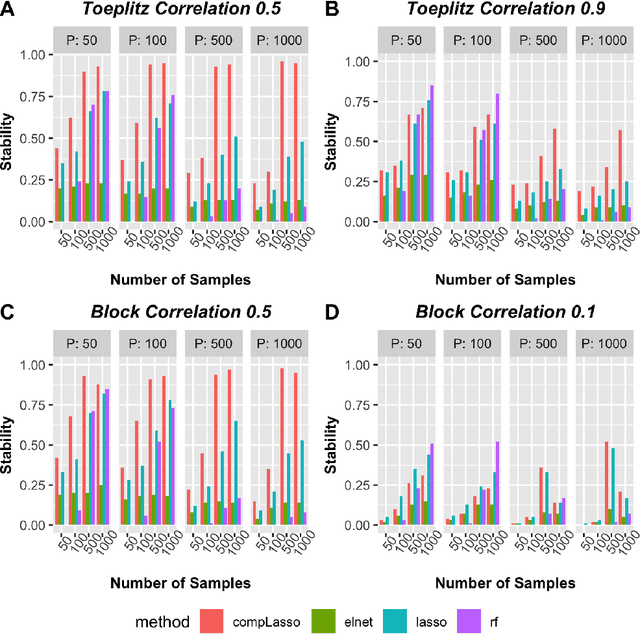
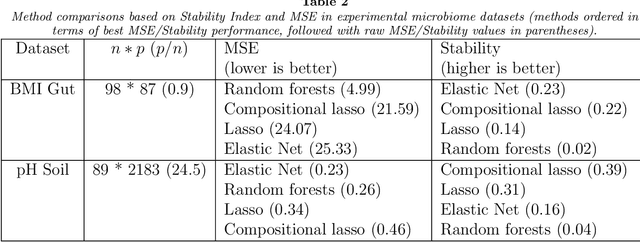
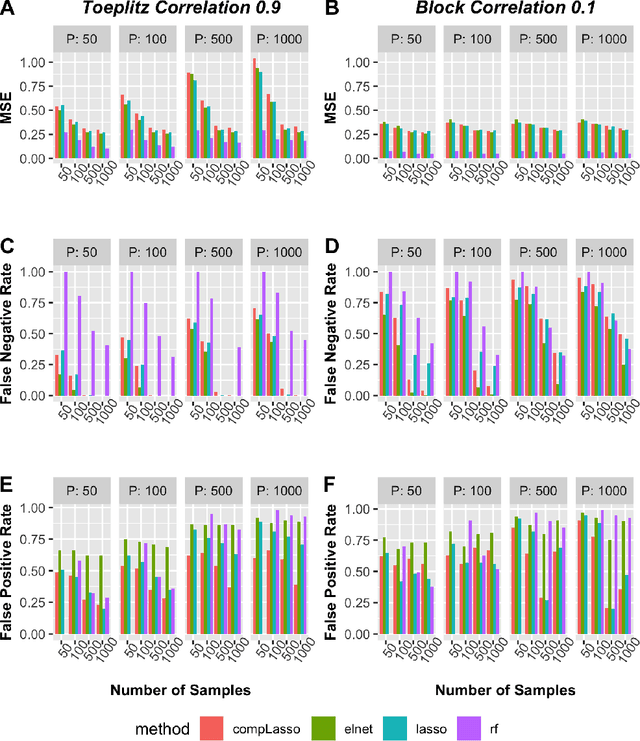
Abstract:Feature selection is indispensable in microbiome data analysis, but it can be particularly challenging as microbiome data sets are high-dimensional, underdetermined, sparse and compositional. Great efforts have recently been made on developing new methods for feature selection that handle the above data characteristics, but almost all methods were evaluated based on performance of model predictions. However, little attention has been paid to address a fundamental question: how appropriate are those evaluation criteria? Most feature selection methods often control the model fit, but the ability to identify meaningful subsets of features cannot be evaluated simply based on the prediction accuracy. If tiny changes to the training data would lead to large changes in the chosen feature subset, then many of the biological features that an algorithm has found are likely to be a data artifact rather than real biological signal. This crucial need of identifying relevant and reproducible features motivated the reproducibility evaluation criterion such as Stability, which quantifies how robust a method is to perturbations in the data. In our paper, we compare the performance of popular model prediction metric MSE and proposed reproducibility criterion Stability in evaluating four widely used feature selection methods in both simulations and experimental microbiome applications. We conclude that Stability is a preferred feature selection criterion over MSE because it better quantifies the reproducibility of the feature selection method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge