Chun-Nan Hsu
Development and external validation of a multimodal artificial intelligence mortality prediction model of critically ill patients using multicenter data
Dec 15, 2025Abstract:Early prediction of in-hospital mortality in critically ill patients can aid clinicians in optimizing treatment. The objective was to develop a multimodal deep learning model, using structured and unstructured clinical data, to predict in-hospital mortality risk among critically ill patients after their initial 24 hour intensive care unit (ICU) admission. We used data from MIMIC-III, MIMIC-IV, eICU, and HiRID. A multimodal model was developed on the MIMIC datasets, featuring time series components occurring within the first 24 hours of ICU admission and predicting risk of subsequent inpatient mortality. Inputs included time-invariant variables, time-variant variables, clinical notes, and chest X-ray images. External validation occurred in a temporally separated MIMIC population, HiRID, and eICU datasets. A total of 203,434 ICU admissions from more than 200 hospitals between 2001 to 2022 were included, in which mortality rate ranged from 5.2% to 7.9% across the four datasets. The model integrating structured data points had AUROC, AUPRC, and Brier scores of 0.92, 0.53, and 0.19, respectively. We externally validated the model on eight different institutions within the eICU dataset, demonstrating AUROCs ranging from 0.84-0.92. When including only patients with available clinical notes and imaging data, inclusion of notes and imaging into the model, the AUROC, AUPRC, and Brier score improved from 0.87 to 0.89, 0.43 to 0.48, and 0.37 to 0.17, respectively. Our findings highlight the importance of incorporating multiple sources of patient information for mortality prediction and the importance of external validation.
MedEval: A Multi-Level, Multi-Task, and Multi-Domain Medical Benchmark for Language Model Evaluation
Oct 27, 2023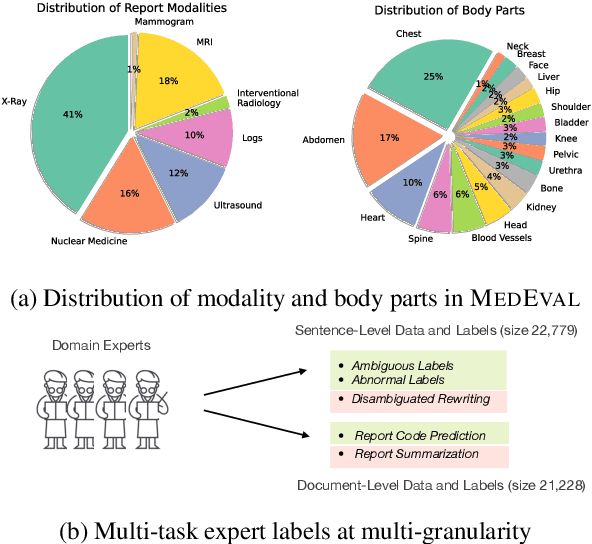
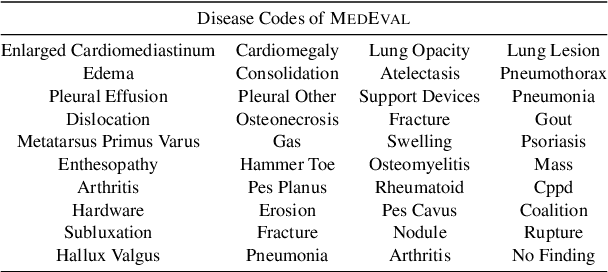
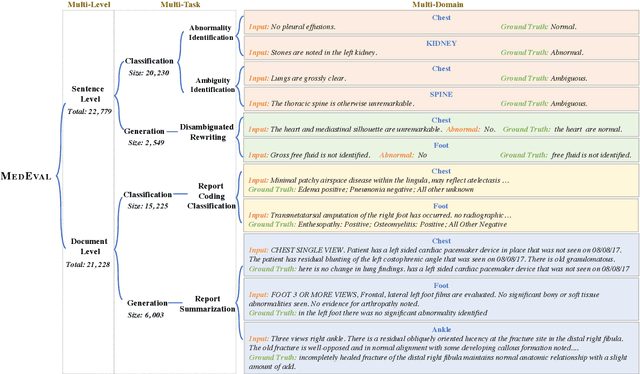
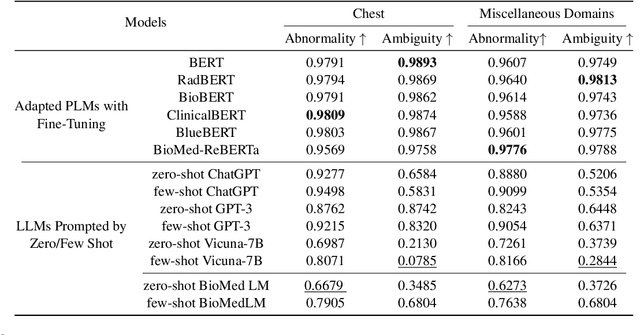
Abstract:Curated datasets for healthcare are often limited due to the need of human annotations from experts. In this paper, we present MedEval, a multi-level, multi-task, and multi-domain medical benchmark to facilitate the development of language models for healthcare. MedEval is comprehensive and consists of data from several healthcare systems and spans 35 human body regions from 8 examination modalities. With 22,779 collected sentences and 21,228 reports, we provide expert annotations at multiple levels, offering a granular potential usage of the data and supporting a wide range of tasks. Moreover, we systematically evaluated 10 generic and domain-specific language models under zero-shot and finetuning settings, from domain-adapted baselines in healthcare to general-purposed state-of-the-art large language models (e.g., ChatGPT). Our evaluations reveal varying effectiveness of the two categories of language models across different tasks, from which we notice the importance of instruction tuning for few-shot usage of large language models. Our investigation paves the way toward benchmarking language models for healthcare and provides valuable insights into the strengths and limitations of adopting large language models in medical domains, informing their practical applications and future advancements.
Robust and Interpretable Medical Image Classifiers via Concept Bottleneck Models
Oct 04, 2023



Abstract:Medical image classification is a critical problem for healthcare, with the potential to alleviate the workload of doctors and facilitate diagnoses of patients. However, two challenges arise when deploying deep learning models to real-world healthcare applications. First, neural models tend to learn spurious correlations instead of desired features, which could fall short when generalizing to new domains (e.g., patients with different ages). Second, these black-box models lack interpretability. When making diagnostic predictions, it is important to understand why a model makes a decision for trustworthy and safety considerations. In this paper, to address these two limitations, we propose a new paradigm to build robust and interpretable medical image classifiers with natural language concepts. Specifically, we first query clinical concepts from GPT-4, then transform latent image features into explicit concepts with a vision-language model. We systematically evaluate our method on eight medical image classification datasets to verify its effectiveness. On challenging datasets with strong confounding factors, our method can mitigate spurious correlations thus substantially outperform standard visual encoders and other baselines. Finally, we show how classification with a small number of concepts brings a level of interpretability for understanding model decisions through case studies in real medical data.
"Nothing Abnormal": Disambiguating Medical Reports via Contrastive Knowledge Infusion
May 15, 2023Abstract:Sharing medical reports is essential for patient-centered care. A recent line of work has focused on automatically generating reports with NLP methods. However, different audiences have different purposes when writing/reading medical reports -- for example, healthcare professionals care more about pathology, whereas patients are more concerned with the diagnosis ("Is there any abnormality?"). The expectation gap results in a common situation where patients find their medical reports to be ambiguous and therefore unsure about the next steps. In this work, we explore the audience expectation gap in healthcare and summarize common ambiguities that lead patients to be confused about their diagnosis into three categories: medical jargon, contradictory findings, and misleading grammatical errors. Based on our analysis, we define a disambiguation rewriting task to regenerate an input to be unambiguous while preserving information about the original content. We further propose a rewriting algorithm based on contrastive pretraining and perturbation-based rewriting. In addition, we create two datasets, OpenI-Annotated based on chest reports and VA-Annotated based on general medical reports, with available binary labels for ambiguity and abnormality presence annotated by radiology specialists. Experimental results on these datasets show that our proposed algorithm effectively rewrites input sentences in a less ambiguous way with high content fidelity. Our code and annotated data are released to facilitate future research.
Representing Knowledge by Spans: A Knowledge-Enhanced Model for Information Extraction
Aug 20, 2022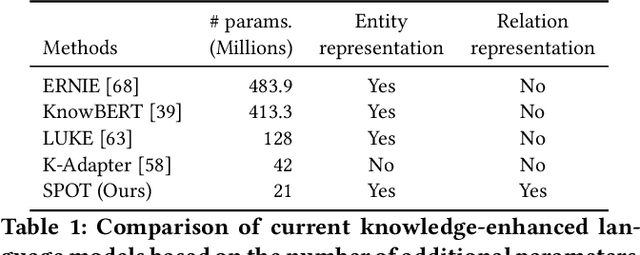
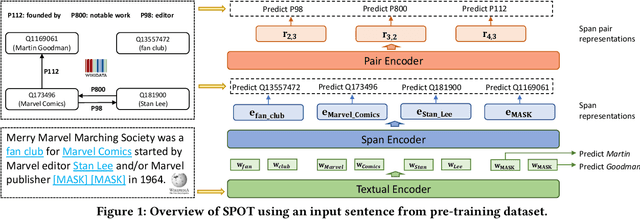
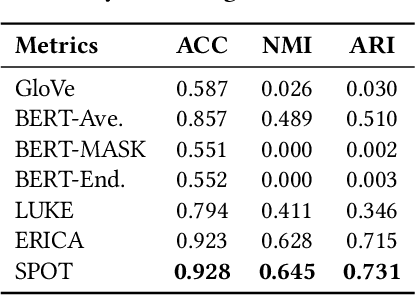
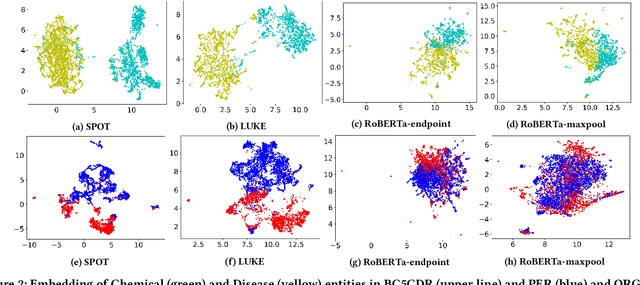
Abstract:Knowledge-enhanced pre-trained models for language representation have been shown to be more effective in knowledge base construction tasks (i.e.,~relation extraction) than language models such as BERT. These knowledge-enhanced language models incorporate knowledge into pre-training to generate representations of entities or relationships. However, existing methods typically represent each entity with a separate embedding. As a result, these methods struggle to represent out-of-vocabulary entities and a large amount of parameters, on top of their underlying token models (i.e.,~the transformer), must be used and the number of entities that can be handled is limited in practice due to memory constraints. Moreover, existing models still struggle to represent entities and relationships simultaneously. To address these problems, we propose a new pre-trained model that learns representations of both entities and relationships from token spans and span pairs in the text respectively. By encoding spans efficiently with span modules, our model can represent both entities and their relationships but requires fewer parameters than existing models. We pre-trained our model with the knowledge graph extracted from Wikipedia and test it on a broad range of supervised and unsupervised information extraction tasks. Results show that our model learns better representations for both entities and relationships than baselines, while in supervised settings, fine-tuning our model outperforms RoBERTa consistently and achieves competitive results on information extraction tasks.
Abstractified Multi-instance Learning (AMIL) for Biomedical Relation Extraction
Oct 24, 2021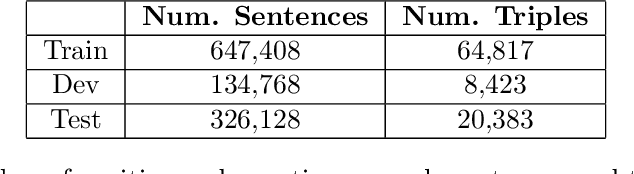

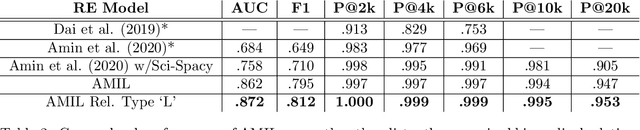
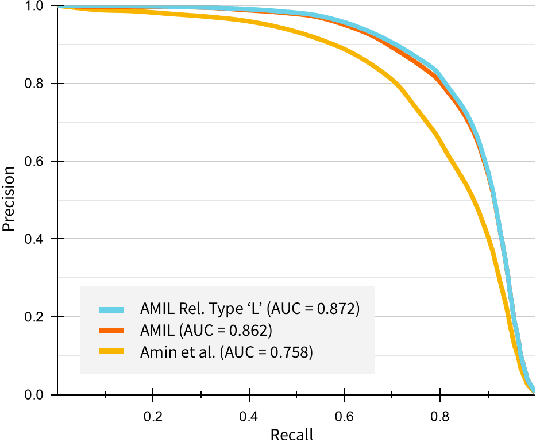
Abstract:Relation extraction in the biomedical domain is a challenging task due to a lack of labeled data and a long-tail distribution of fact triples. Many works leverage distant supervision which automatically generates labeled data by pairing a knowledge graph with raw textual data. Distant supervision produces noisy labels and requires additional techniques, such as multi-instance learning (MIL), to denoise the training signal. However, MIL requires multiple instances of data and struggles with very long-tail datasets such as those found in the biomedical domain. In this work, we propose a novel reformulation of MIL for biomedical relation extraction that abstractifies biomedical entities into their corresponding semantic types. By grouping entities by types, we are better able to take advantage of the benefits of MIL and further denoise the training signal. We show this reformulation, which we refer to as abstractified multi-instance learning (AMIL), improves performance in biomedical relationship extraction. We also propose a novel relationship embedding architecture that further improves model performance.
* 14 pages, 3 figures, submitted to Automated Knowledge Base Construction (2021)
Weakly Supervised Contrastive Learning for Chest X-Ray Report Generation
Sep 25, 2021



Abstract:Radiology report generation aims at generating descriptive text from radiology images automatically, which may present an opportunity to improve radiology reporting and interpretation. A typical setting consists of training encoder-decoder models on image-report pairs with a cross entropy loss, which struggles to generate informative sentences for clinical diagnoses since normal findings dominate the datasets. To tackle this challenge and encourage more clinically-accurate text outputs, we propose a novel weakly supervised contrastive loss for medical report generation. Experimental results demonstrate that our method benefits from contrasting target reports with incorrect but semantically-close ones. It outperforms previous work on both clinical correctness and text generation metrics for two public benchmarks.
Theoretical Knowledge Graph Reasoning via Ending Anchored Rules
Dec 15, 2020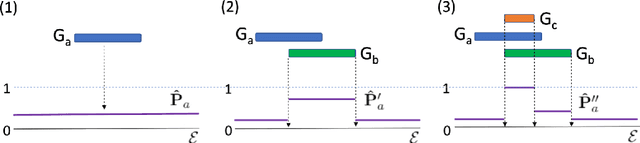

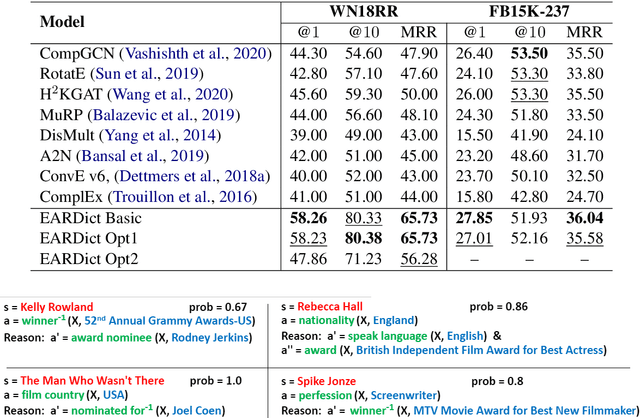
Abstract:Discovering precise and specific rules from knowledge graphs is regarded as an essential challenge, which can improve the performances of many downstream tasks and even provide new ways to approach some Natural Language Processing research topics. In this paper, we provide a fundamental theory for knowledge graph reasoning based on the ending anchored rules. Our theory provides precise reasons explaining why or why not a triple is correct. Then, we implement our theory by what we call the EARDict model. Results show that our EARDict model significantly outperforms all the benchmark models on two large datasets of knowledge graph completion, including achieving a Hits@10 score of 96.6 percent on WN18RR.
Learning Visual-Semantic Embeddings for Reporting Abnormal Findings on Chest X-rays
Oct 06, 2020



Abstract:Automatic medical image report generation has drawn growing attention due to its potential to alleviate radiologists' workload. Existing work on report generation often trains encoder-decoder networks to generate complete reports. However, such models are affected by data bias (e.g.~label imbalance) and face common issues inherent in text generation models (e.g.~repetition). In this work, we focus on reporting abnormal findings on radiology images; instead of training on complete radiology reports, we propose a method to identify abnormal findings from the reports in addition to grouping them with unsupervised clustering and minimal rules. We formulate the task as cross-modal retrieval and propose Conditional Visual-Semantic Embeddings to align images and fine-grained abnormal findings in a joint embedding space. We demonstrate that our method is able to retrieve abnormal findings and outperforms existing generation models on both clinical correctness and text generation metrics.
Antibody Watch: Text Mining Antibody Specificity from the Literature
Aug 05, 2020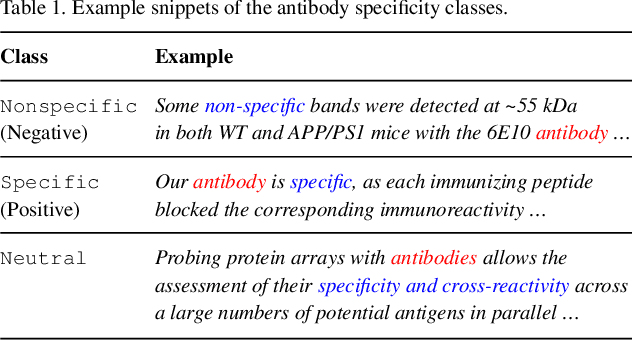

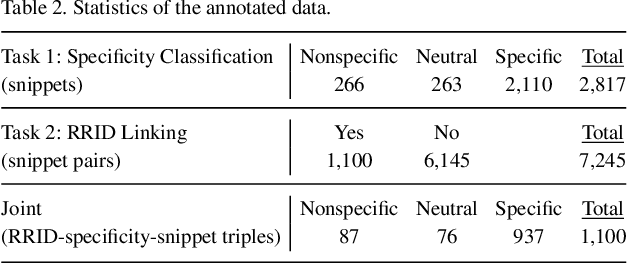
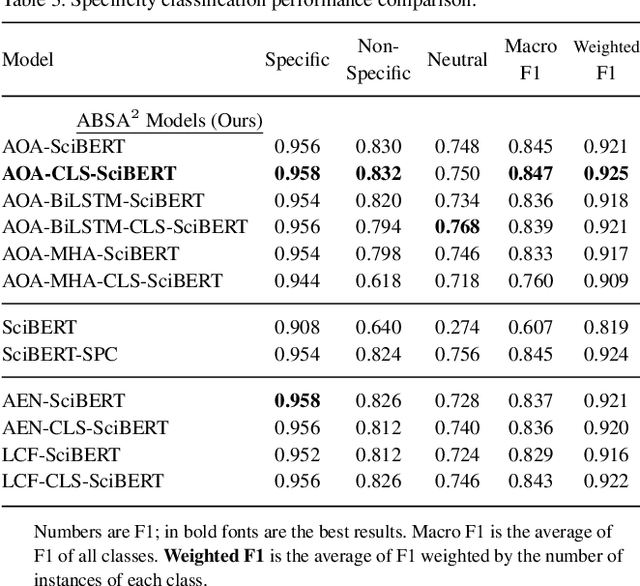
Abstract:Motivation: Antibodies are widely used reagents to test for expression of proteins. However, they might not always reliably produce results when they do not specifically bind to the target proteins that their providers designed them for, leading to unreliable research results. While many proposals have been developed to deal with the problem of antibody specificity, they may not scale well to deal with the millions of antibodies that are available to researchers. In this study, we investigate the feasibility of automatically generating a report to alert users of problematic antibodies by extracting statements about antibody specificity reported in the literature. Results: Our goal is to construct an "Antibody Watch" knowledge base containing supporting statements of problematic antibodies. We developed a deep neural network system and tested its performance with a corpus of more than two thousand articles that reported uses of antibodies. We divided the problem into two tasks. Given an input article, the first task is to identify snippets about antibody specificity and classify if the snippets report that any antibody exhibits nonspecificity, and thus is problematic. The second task is to link each of these snippets to one or more antibodies mentioned in the snippet. The experimental evaluation shows that our system can accurately perform both classification and linking tasks with weighted F-scores over 0.925 and 0.923, respectively, and 0.914 overall when combined to complete the joint task. We leveraged Research Resource Identifiers (RRID) to precisely identify antibodies linked to the extracted specificity snippets. The result shows that it is feasible to construct a reliable knowledge base about problematic antibodies by text mining.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge