Lucila Ohno-Machado
UC San Diego
A Federated and Parameter-Efficient Framework for Large Language Model Training in Medicine
Jan 29, 2026Abstract:Large language models (LLMs) have demonstrated strong performance on medical benchmarks, including question answering and diagnosis. To enable their use in clinical settings, LLMs are typically further adapted through continued pretraining or post-training using clinical data. However, most medical LLMs are trained on data from a single institution, which faces limitations in generalizability and safety in heterogeneous systems. Federated learning (FL) is a promising solution for enabling collaborative model development across healthcare institutions. Yet applying FL to LLMs in medicine remains fundamentally limited. First, conventional FL requires transmitting the full model during each communication round, which becomes impractical for multi-billion-parameter LLMs given the limited computational resources. Second, many FL algorithms implicitly assume data homogeneity, whereas real-world clinical data are highly heterogeneous across patients, diseases, and institutional practices. We introduce the model-agnostic and parameter-efficient federated learning framework for adapting LLMs to medical applications. Fed-MedLoRA transmits only low-rank adapter parameters, reducing communication and computation overhead, while Fed-MedLoRA+ further incorporates adaptive, data-aware aggregation to improve convergence under cross-site heterogeneity. We apply the framework to clinical information extraction (IE), which transforms patient narratives into structured medical entities and relations. Accuracy was assessed across five patient cohorts through comparisons with BERT models, and LLaMA-3 and DeepSeek-R1, GPT-4o models. Evaluation settings included (1) in-domain training and testing, (2) external validation on independent cohorts, and (3) a low-resource new-site adaptation scenario using real-world clinical notes from the Yale New Haven Health System.
Near-Lossless Model Compression Enables Longer Context Inference in DNA Large Language Models
Nov 18, 2025Abstract:Trained on massive cross-species DNA corpora, DNA large language models (LLMs) learn the fundamental "grammar" and evolutionary patterns of genomic sequences. This makes them powerful priors for DNA sequence modeling, particularly over long ranges. However, two major constraints hinder their use in practice: the quadratic computational cost of self-attention and the growing memory required for key-value (KV) caches during autoregressive decoding. These constraints force the use of heuristics such as fixed-window truncation or sliding windows, which compromise fidelity on ultra-long sequences by discarding distant information. We introduce FOCUS (Feature-Oriented Compression for Ultra-long Self-attention), a progressive context-compression module that can be plugged into pretrained DNA LLMs. FOCUS combines the established k-mer representation in genomics with learnable hierarchical compression: it inserts summary tokens at k-mer granularity and progressively compresses attention key and value activations across multiple Transformer layers, retaining only the summary KV states across windows while discarding ordinary-token KV. A shared-boundary windowing scheme yields a stationary cross-window interface that propagates long-range information with minimal loss. We validate FOCUS on an Evo-2-based DNA LLM fine-tuned on GRCh38 chromosome 1 with self-supervised training and randomized compression schedules to promote robustness across compression ratios. On held-out human chromosomes, FOCUS achieves near-lossless fidelity: compressing a 1 kb context into only 10 summary tokens (about 100x) shifts the average per-nucleotide probability by only about 0.0004. Compared to a baseline without compression, FOCUS reduces KV-cache memory and converts effective inference scaling from O(N^2) to near-linear O(N), enabling about 100x longer inference windows on commodity GPUs with near-lossless fidelity.
Memorization in Large Language Models in Medicine: Prevalence, Characteristics, and Implications
Sep 10, 2025Abstract:Large Language Models (LLMs) have demonstrated significant potential in medicine. To date, LLMs have been widely applied to tasks such as diagnostic assistance, medical question answering, and clinical information synthesis. However, a key open question remains: to what extent do LLMs memorize medical training data. In this study, we present the first comprehensive evaluation of memorization of LLMs in medicine, assessing its prevalence (how frequently it occurs), characteristics (what is memorized), volume (how much content is memorized), and potential downstream impacts (how memorization may affect medical applications). We systematically analyze common adaptation scenarios: (1) continued pretraining on medical corpora, (2) fine-tuning on standard medical benchmarks, and (3) fine-tuning on real-world clinical data, including over 13,000 unique inpatient records from Yale New Haven Health System. The results demonstrate that memorization is prevalent across all adaptation scenarios and significantly higher than reported in the general domain. Memorization affects both the development and adoption of LLMs in medicine and can be categorized into three types: beneficial (e.g., accurate recall of clinical guidelines and biomedical references), uninformative (e.g., repeated disclaimers or templated medical document language), and harmful (e.g., regeneration of dataset-specific or sensitive clinical content). Based on these findings, we offer practical recommendations to facilitate beneficial memorization that enhances domain-specific reasoning and factual accuracy, minimize uninformative memorization to promote deeper learning beyond surface-level patterns, and mitigate harmful memorization to prevent the leakage of sensitive or identifiable patient information.
Hey, That's My Data! Label-Only Dataset Inference in Large Language Models
Jun 06, 2025Abstract:Large Language Models (LLMs) have revolutionized Natural Language Processing by excelling at interpreting, reasoning about, and generating human language. However, their reliance on large-scale, often proprietary datasets poses a critical challenge: unauthorized usage of such data can lead to copyright infringement and significant financial harm. Existing dataset-inference methods typically depend on log probabilities to detect suspicious training material, yet many leading LLMs have begun withholding or obfuscating these signals. This reality underscores the pressing need for label-only approaches capable of identifying dataset membership without relying on internal model logits. We address this gap by introducing CatShift, a label-only dataset-inference framework that capitalizes on catastrophic forgetting: the tendency of an LLM to overwrite previously learned knowledge when exposed to new data. If a suspicious dataset was previously seen by the model, fine-tuning on a portion of it triggers a pronounced post-tuning shift in the model's outputs; conversely, truly novel data elicits more modest changes. By comparing the model's output shifts for a suspicious dataset against those for a known non-member validation set, we statistically determine whether the suspicious set is likely to have been part of the model's original training corpus. Extensive experiments on both open-source and API-based LLMs validate CatShift's effectiveness in logit-inaccessible settings, offering a robust and practical solution for safeguarding proprietary data.
Enhancing Patient-Centric Communication: Leveraging LLMs to Simulate Patient Perspectives
Jan 12, 2025



Abstract:Large Language Models (LLMs) have demonstrated impressive capabilities in role-playing scenarios, particularly in simulating domain-specific experts using tailored prompts. This ability enables LLMs to adopt the persona of individuals with specific backgrounds, offering a cost-effective and efficient alternative to traditional, resource-intensive user studies. By mimicking human behavior, LLMs can anticipate responses based on concrete demographic or professional profiles. In this paper, we evaluate the effectiveness of LLMs in simulating individuals with diverse backgrounds and analyze the consistency of these simulated behaviors compared to real-world outcomes. In particular, we explore the potential of LLMs to interpret and respond to discharge summaries provided to patients leaving the Intensive Care Unit (ICU). We evaluate and compare with human responses the comprehensibility of discharge summaries among individuals with varying educational backgrounds, using this analysis to assess the strengths and limitations of LLM-driven simulations. Notably, when LLMs are primed with educational background information, they deliver accurate and actionable medical guidance 88% of the time. However, when other information is provided, performance significantly drops, falling below random chance levels. This preliminary study shows the potential benefits and pitfalls of automatically generating patient-specific health information from diverse populations. While LLMs show promise in simulating health personas, our results highlight critical gaps that must be addressed before they can be reliably used in clinical settings. Our findings suggest that a straightforward query-response model could outperform a more tailored approach in delivering health information. This is a crucial first step in understanding how LLMs can be optimized for personalized health communication while maintaining accuracy.
The Impact of Automatic Pre-annotation in Clinical Note Data Element Extraction - the CLEAN Tool
Aug 11, 2018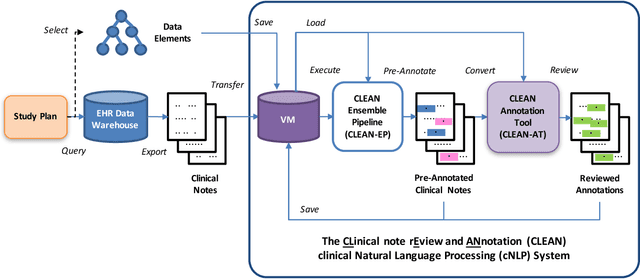
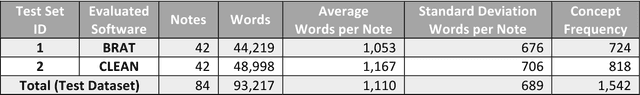
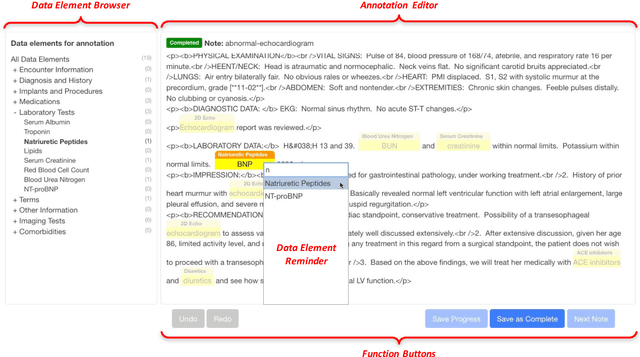
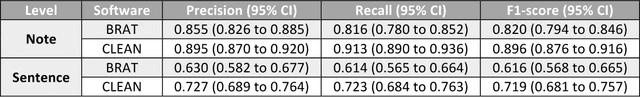
Abstract:Objective. Annotation is expensive but essential for clinical note review and clinical natural language processing (cNLP). However, the extent to which computer-generated pre-annotation is beneficial to human annotation is still an open question. Our study introduces CLEAN (CLinical note rEview and ANnotation), a pre-annotation-based cNLP annotation system to improve clinical note annotation of data elements, and comprehensively compares CLEAN with the widely-used annotation system Brat Rapid Annotation Tool (BRAT). Materials and Methods. CLEAN includes an ensemble pipeline (CLEAN-EP) with a newly developed annotation tool (CLEAN-AT). A domain expert and a novice user/annotator participated in a comparative usability test by tagging 87 data elements related to Congestive Heart Failure (CHF) and Kawasaki Disease (KD) cohorts in 84 public notes. Results. CLEAN achieved higher note-level F1-score (0.896) over BRAT (0.820), with significant difference in correctness (P-value < 0.001), and the mostly related factor being system/software (P-value < 0.001). No significant difference (P-value 0.188) in annotation time was observed between CLEAN (7.262 minutes/note) and BRAT (8.286 minutes/note). The difference was mostly associated with note length (P-value < 0.001) and system/software (P-value 0.013). The expert reported CLEAN to be useful/satisfactory, while the novice reported slight improvements. Discussion. CLEAN improves the correctness of annotation and increases usefulness/satisfaction with the same level of efficiency. Limitations include untested impact of pre-annotation correctness rate, small sample size, small user size, and restrictedly validated gold standard. Conclusion. CLEAN with pre-annotation can be beneficial for an expert to deal with complex annotation tasks involving numerous and diverse target data elements.
Natural Language Processing in Biomedicine: A Unified System Architecture Overview
Jan 08, 2014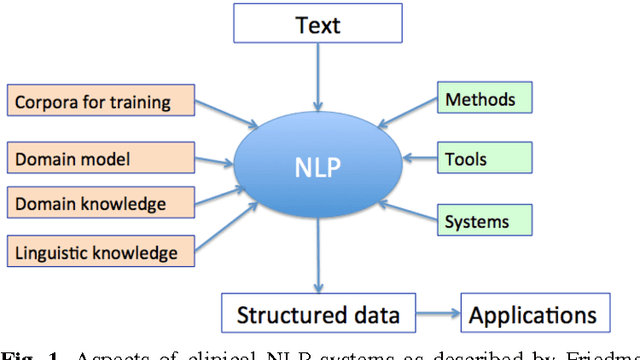
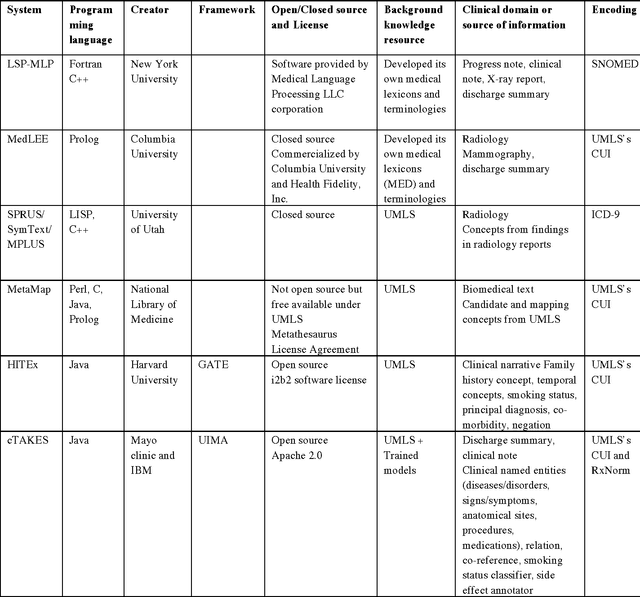
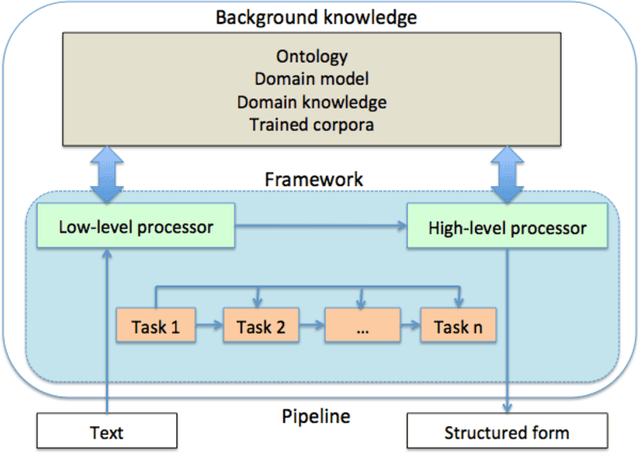
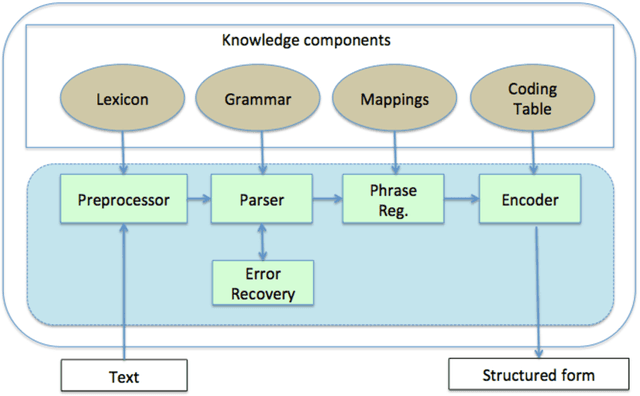
Abstract:In modern electronic medical records (EMR) much of the clinically important data - signs and symptoms, symptom severity, disease status, etc. - are not provided in structured data fields, but rather are encoded in clinician generated narrative text. Natural language processing (NLP) provides a means of "unlocking" this important data source for applications in clinical decision support, quality assurance, and public health. This chapter provides an overview of representative NLP systems in biomedicine based on a unified architectural view. A general architecture in an NLP system consists of two main components: background knowledge that includes biomedical knowledge resources and a framework that integrates NLP tools to process text. Systems differ in both components, which we will review briefly. Additionally, challenges facing current research efforts in biomedical NLP include the paucity of large, publicly available annotated corpora, although initiatives that facilitate data sharing, system evaluation, and collaborative work between researchers in clinical NLP are starting to emerge.
Enhancing Twitter Data Analysis with Simple Semantic Filtering: Example in Tracking Influenza-Like Illnesses
Oct 02, 2012



Abstract:Systems that exploit publicly available user generated content such as Twitter messages have been successful in tracking seasonal influenza. We developed a novel filtering method for Influenza-Like-Illnesses (ILI)-related messages using 587 million messages from Twitter micro-blogs. We first filtered messages based on syndrome keywords from the BioCaster Ontology, an extant knowledge model of laymen's terms. We then filtered the messages according to semantic features such as negation, hashtags, emoticons, humor and geography. The data covered 36 weeks for the US 2009 influenza season from 30th August 2009 to 8th May 2010. Results showed that our system achieved the highest Pearson correlation coefficient of 98.46% (p-value<2.2e-16), an improvement of 3.98% over the previous state-of-the-art method. The results indicate that simple NLP-based enhancements to existing approaches to mine Twitter data can increase the value of this inexpensive resource.
Predicting accurate probabilities with a ranking loss
Jun 18, 2012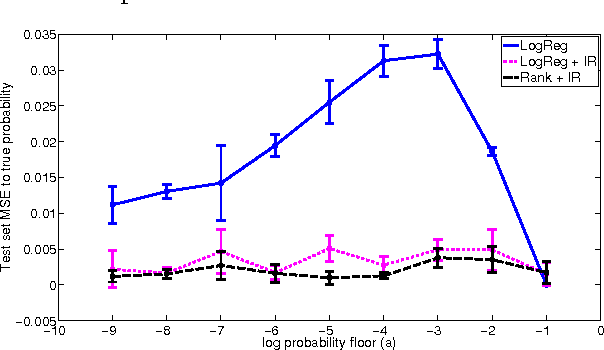
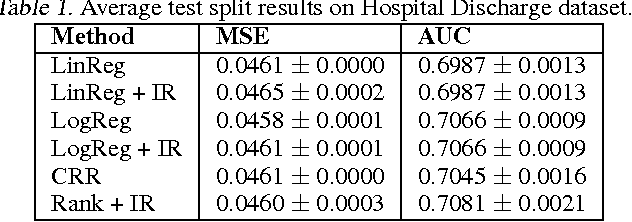
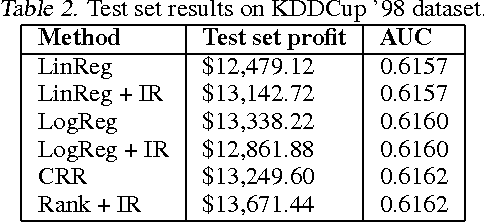
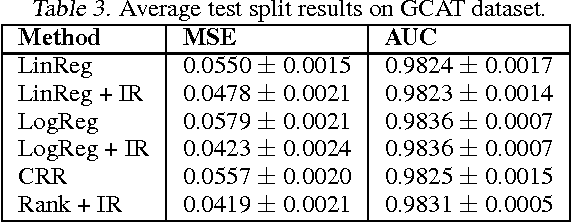
Abstract:In many real-world applications of machine learning classifiers, it is essential to predict the probability of an example belonging to a particular class. This paper proposes a simple technique for predicting probabilities based on optimizing a ranking loss, followed by isotonic regression. This semi-parametric technique offers both good ranking and regression performance, and models a richer set of probability distributions than statistical workhorses such as logistic regression. We provide experimental results that show the effectiveness of this technique on real-world applications of probability prediction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge