John Levander
Creating a Discipline-specific Commons for Infectious Disease Epidemiology
Nov 12, 2023Abstract:Objective: To create a commons for infectious disease (ID) epidemiology in which epidemiologists, public health officers, data producers, and software developers can not only share data and software, but receive assistance in improving their interoperability. Materials and Methods: We represented 586 datasets, 54 software, and 24 data formats in OWL 2 and then used logical queries to infer potentially interoperable combinations of software and datasets, as well as statistics about the FAIRness of the collection. We represented the objects in DATS 2.2 and a software metadata schema of our own design. We used these representations as the basis for the Content, Search, FAIR-o-meter, and Workflow pages that constitute the MIDAS Digital Commons. Results: Interoperability was limited by lack of standardization of input and output formats of software. When formats existed, they were human-readable specifications (22/24; 92%); only 3 formats (13%) had machine-readable specifications. Nevertheless, logical search of a triple store based on named data formats was able to identify scores of potentially interoperable combinations of software and datasets. Discussion: We improved the findability and availability of a sample of software and datasets and developed metrics for assessing interoperability. The barriers to interoperability included poor documentation of software input/output formats and little attention to standardization of most types of data in this field. Conclusion: Centralizing and formalizing the representation of digital objects within a commons promotes FAIRness, enables its measurement over time and the identification of potentially interoperable combinations of data and software.
Bayesian Biosurveillance of Disease Outbreaks
Jul 11, 2012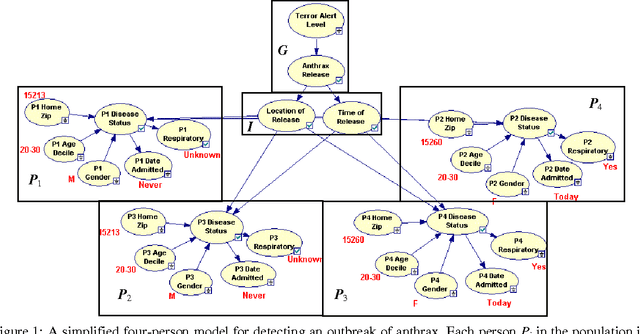
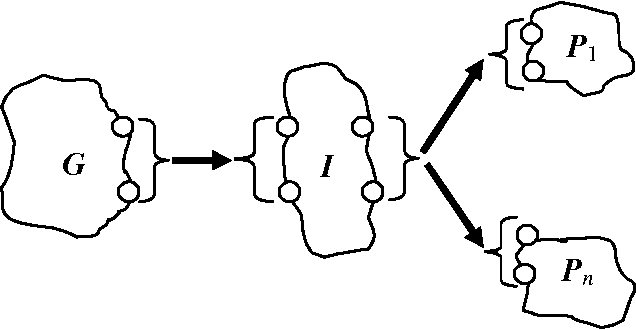
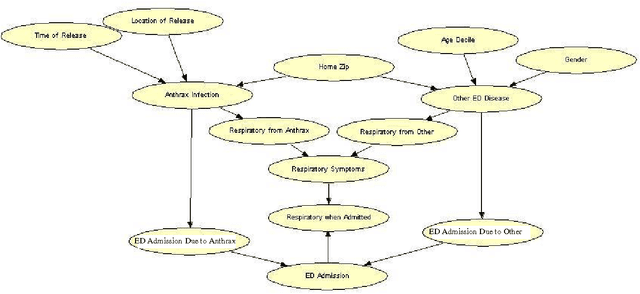
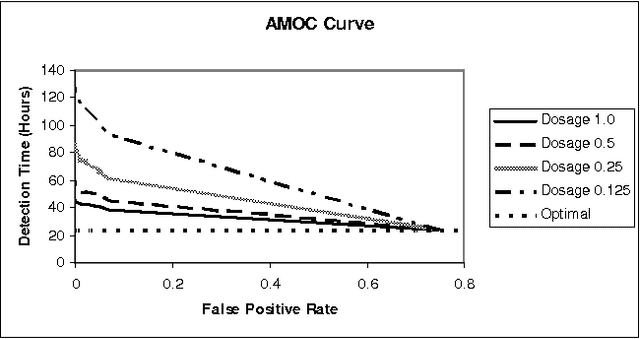
Abstract:Early, reliable detection of disease outbreaks is a critical problem today. This paper reports an investigation of the use of causal Bayesian networks to model spatio-temporal patterns of a non-contagious disease (respiratory anthrax infection) in a population of people. The number of parameters in such a network can become enormous, if not carefully managed. Also, inference needs to be performed in real time as population data stream in. We describe techniques we have applied to address both the modeling and inference challenges. A key contribution of this paper is the explication of assumptions and techniques that are sufficient to allow the scaling of Bayesian network modeling and inference to millions of nodes for real-time surveillance applications. The results reported here provide a proof-of-concept that Bayesian networks can serve as the foundation of a system that effectively performs Bayesian biosurveillance of disease outbreaks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge