Hamid Alinejad-Rokny
Beyond Quantity: Trajectory Diversity Scaling for Code Agents
Feb 03, 2026Abstract:As code large language models (LLMs) evolve into tool-interactive agents via the Model Context Protocol (MCP), their generalization is increasingly limited by low-quality synthetic data and the diminishing returns of quantity scaling. Moreover, quantity-centric scaling exhibits an early bottleneck that underutilizes trajectory data. We propose TDScaling, a Trajectory Diversity Scaling-based data synthesis framework for code agents that scales performance through diversity rather than raw volume. Under a fixed training budget, increasing trajectory diversity yields larger gains than adding more trajectories, improving the performance-cost trade-off for agent training. TDScaling integrates four innovations: (1) a Business Cluster mechanism that captures real-service logical dependencies; (2) a blueprint-driven multi-agent paradigm that enforces trajectory coherence; (3) an adaptive evolution mechanism that steers synthesis toward long-tail scenarios using Domain Entropy, Reasoning Mode Entropy, and Cumulative Action Complexity to prevent mode collapse; and (4) a sandboxed code tool that mitigates catastrophic forgetting of intrinsic coding capabilities. Experiments on general tool-use benchmarks (BFCL, tau^2-Bench) and code agent tasks (RebenchT, CodeCI, BIRD) demonstrate a win-win outcome: TDScaling improves both tool-use generalization and inherent coding proficiency. We plan to release the full codebase and the synthesized dataset (including 30,000+ tool clusters) upon publication.
Automatic Paper Reviewing with Heterogeneous Graph Reasoning over LLM-Simulated Reviewer-Author Debates
Nov 11, 2025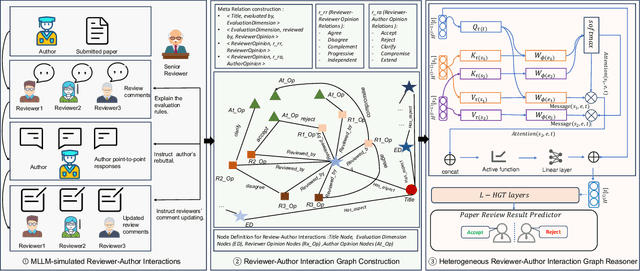
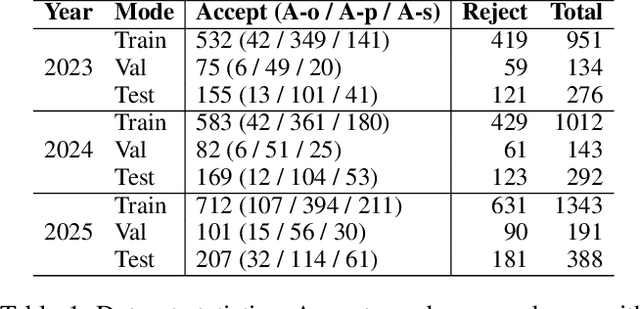
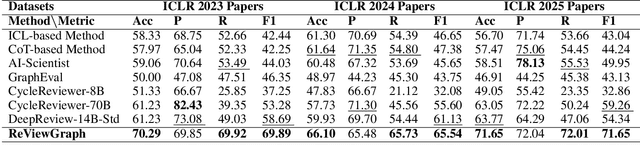
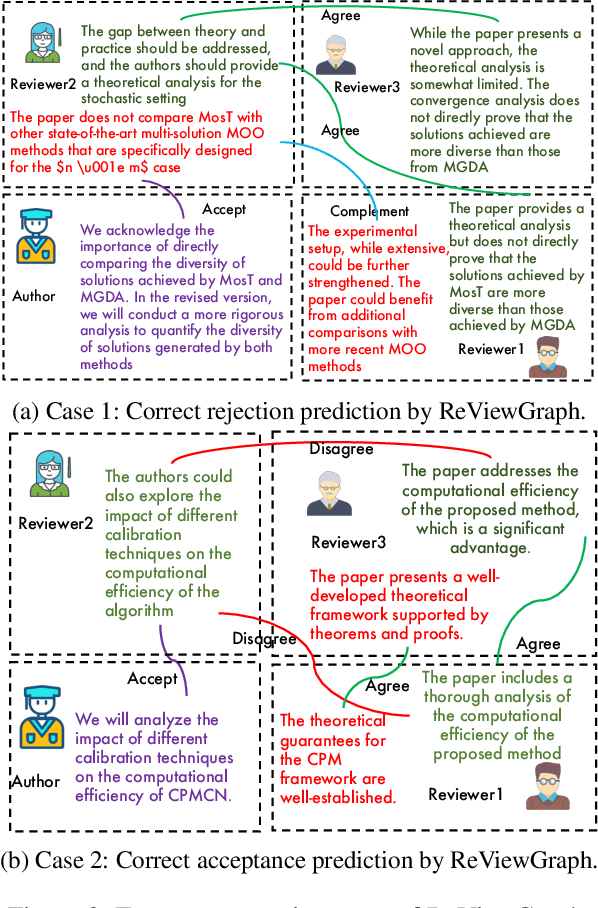
Abstract:Existing paper review methods often rely on superficial manuscript features or directly on large language models (LLMs), which are prone to hallucinations, biased scoring, and limited reasoning capabilities. Moreover, these methods often fail to capture the complex argumentative reasoning and negotiation dynamics inherent in reviewer-author interactions. To address these limitations, we propose ReViewGraph (Reviewer-Author Debates Graph Reasoner), a novel framework that performs heterogeneous graph reasoning over LLM-simulated multi-round reviewer-author debates. In our approach, reviewer-author exchanges are simulated through LLM-based multi-agent collaboration. Diverse opinion relations (e.g., acceptance, rejection, clarification, and compromise) are then explicitly extracted and encoded as typed edges within a heterogeneous interaction graph. By applying graph neural networks to reason over these structured debate graphs, ReViewGraph captures fine-grained argumentative dynamics and enables more informed review decisions. Extensive experiments on three datasets demonstrate that ReViewGraph outperforms strong baselines with an average relative improvement of 15.73%, underscoring the value of modeling detailed reviewer-author debate structures.
RxSafeBench: Identifying Medication Safety Issues of Large Language Models in Simulated Consultation
Nov 06, 2025Abstract:Numerous medical systems powered by Large Language Models (LLMs) have achieved remarkable progress in diverse healthcare tasks. However, research on their medication safety remains limited due to the lack of real world datasets, constrained by privacy and accessibility issues. Moreover, evaluation of LLMs in realistic clinical consultation settings, particularly regarding medication safety, is still underexplored. To address these gaps, we propose a framework that simulates and evaluates clinical consultations to systematically assess the medication safety capabilities of LLMs. Within this framework, we generate inquiry diagnosis dialogues with embedded medication risks and construct a dedicated medication safety database, RxRisk DB, containing 6,725 contraindications, 28,781 drug interactions, and 14,906 indication-drug pairs. A two-stage filtering strategy ensures clinical realism and professional quality, resulting in the benchmark RxSafeBench with 2,443 high-quality consultation scenarios. We evaluate leading open-source and proprietary LLMs using structured multiple choice questions that test their ability to recommend safe medications under simulated patient contexts. Results show that current LLMs struggle to integrate contraindication and interaction knowledge, especially when risks are implied rather than explicit. Our findings highlight key challenges in ensuring medication safety in LLM-based systems and provide insights into improving reliability through better prompting and task-specific tuning. RxSafeBench offers the first comprehensive benchmark for evaluating medication safety in LLMs, advancing safer and more trustworthy AI-driven clinical decision support.
One Model to Critique Them All: Rewarding Agentic Tool-Use via Efficient Reasoning
Oct 30, 2025

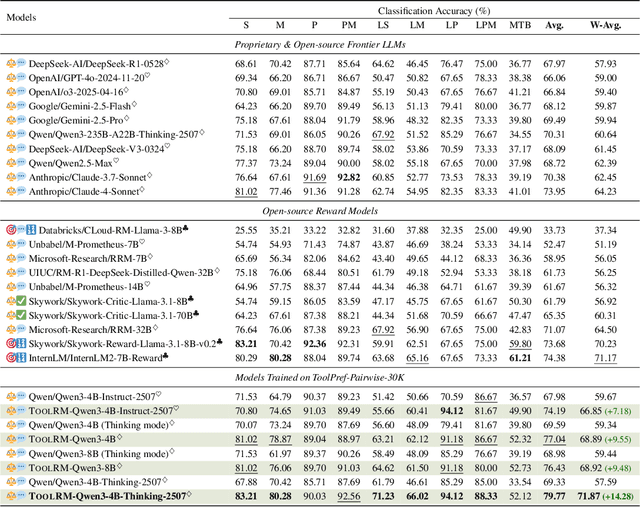

Abstract:Reward models (RMs) play a critical role in aligning large language models (LLMs) with human preferences. Yet in the domain of tool learning, the lack of RMs specifically designed for function-calling tasks has limited progress toward more capable agentic AI. We introduce ToolRM, a family of lightweight generative RMs tailored for general tool-use scenarios. To build these models, we propose a novel pipeline that constructs pairwise preference data using rule-based scoring and multidimensional sampling. This yields ToolPref-Pairwise-30K, a diverse, balanced, and challenging dataset of critique tasks that supports reinforcement learning with verifiable feedback. To evaluate tool-use RMs, we also introduce TRBench$_{BFCL}$, a benchmark built on the agentic evaluation suite BFCL. Trained on our constructed data, models from the Qwen3-4B/8B series achieve up to 14.28% higher accuracy, substantially outperforming frontier models such as Claude 4 and OpenAI o3 in pairwise reward judgments. Beyond training objectives, ToolRM generalizes to broader critique tasks, including Best-of-N sampling and self-correction. Experiments on ACEBench highlight its effectiveness and efficiency, enabling inference-time scaling and reducing output token usage by over 66%. We release data and model checkpoints to facilitate future research.
SemanticST: Spatially Informed Semantic Graph Learning for Clustering, Integration, and Scalable Analysis of Spatial Transcriptomics
Jun 16, 2025Abstract:Spatial transcriptomics (ST) technologies enable gene expression profiling with spatial resolution, offering unprecedented insights into tissue organization and disease heterogeneity. However, current analysis methods often struggle with noisy data, limited scalability, and inadequate modelling of complex cellular relationships. We present SemanticST, a biologically informed, graph-based deep learning framework that models diverse cellular contexts through multi-semantic graph construction. SemanticST builds multiple context-specific graphs capturing spatial proximity, gene expression similarity, and tissue domain structure, and learns disentangled embeddings for each. These are fused using an attention-inspired strategy to yield a unified, biologically meaningful representation. A community-aware min-cut loss improves robustness over contrastive learning, particularly in sparse ST data. SemanticST supports mini-batch training, making it the first graph neural network scalable to large-scale datasets such as Xenium (500,000 cells). Benchmarking across four platforms (Visium, Slide-seq, Stereo-seq, Xenium) and multiple human and mouse tissues shows consistent 20 percentage gains in ARI, NMI, and trajectory fidelity over DeepST, GraphST, and IRIS. In re-analysis of breast cancer Xenium data, SemanticST revealed rare and clinically significant niches, including triple receptor-positive clusters, spatially distinct DCIS-to-IDC transition zones, and FOXC2 tumour-associated myoepithelial cells, suggesting non-canonical EMT programs with stem-like features. SemanticST thus provides a scalable, interpretable, and biologically grounded framework for spatial transcriptomics analysis, enabling robust discovery across tissue types and diseases, and paving the way for spatially resolved tissue atlases and next-generation precision medicine.
Training Superior Sparse Autoencoders for Instruct Models
Jun 09, 2025Abstract:As large language models (LLMs) grow in scale and capability, understanding their internal mechanisms becomes increasingly critical. Sparse autoencoders (SAEs) have emerged as a key tool in mechanistic interpretability, enabling the extraction of human-interpretable features from LLMs. However, existing SAE training methods are primarily designed for base models, resulting in reduced reconstruction quality and interpretability when applied to instruct models. To bridge this gap, we propose $\underline{\textbf{F}}$inetuning-$\underline{\textbf{a}}$ligned $\underline{\textbf{S}}$equential $\underline{\textbf{T}}$raining ($\textit{FAST}$), a novel training method specifically tailored for instruct models. $\textit{FAST}$ aligns the training process with the data distribution and activation patterns characteristic of instruct models, resulting in substantial improvements in both reconstruction and feature interpretability. On Qwen2.5-7B-Instruct, $\textit{FAST}$ achieves a mean squared error of 0.6468 in token reconstruction, significantly outperforming baseline methods with errors of 5.1985 and 1.5096. In feature interpretability, $\textit{FAST}$ yields a higher proportion of high-quality features, for Llama3.2-3B-Instruct, $21.1\%$ scored in the top range, compared to $7.0\%$ and $10.2\%$ for $\textit{BT(P)}$ and $\textit{BT(F)}$. Surprisingly, we discover that intervening on the activations of special tokens via the SAEs leads to improvements in output quality, suggesting new opportunities for fine-grained control of model behavior. Code, data, and 240 trained SAEs are available at https://github.com/Geaming2002/FAST.
CLaSp: In-Context Layer Skip for Self-Speculative Decoding
May 30, 2025Abstract:Speculative decoding (SD) is a promising method for accelerating the decoding process of Large Language Models (LLMs). The efficiency of SD primarily hinges on the consistency between the draft model and the verify model. However, existing drafting approaches typically require additional modules to be trained, which can be challenging to implement and ensure compatibility across various LLMs. In this paper, we propose CLaSp, an in-context layer-skipping strategy for self-speculative decoding. Unlike prior methods, CLaSp does not require additional drafting modules or extra training. Instead, it employs a plug-and-play mechanism by skipping intermediate layers of the verify model to construct a compressed draft model. Specifically, we develop a dynamic programming algorithm that optimizes the layer-skipping process by leveraging the complete hidden states from the last verification stage as an objective. This enables CLaSp to dynamically adjust its layer-skipping strategy after each verification stage, without relying on pre-optimized sets of skipped layers. Experimental results across diverse downstream tasks demonstrate that CLaSp achieves a speedup of 1.3x ~ 1.7x on LLaMA3 series models without altering the original distribution of the generated text.
ChARM: Character-based Act-adaptive Reward Modeling for Advanced Role-Playing Language Agents
May 29, 2025
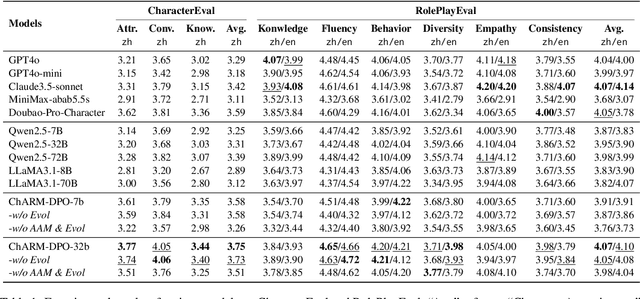
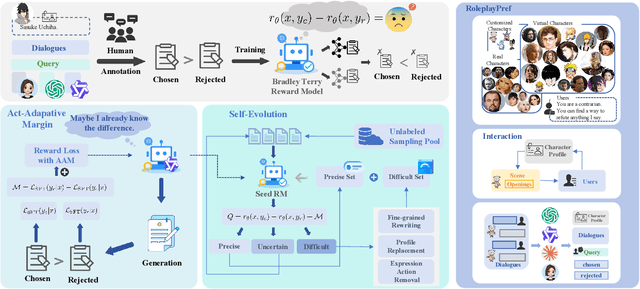
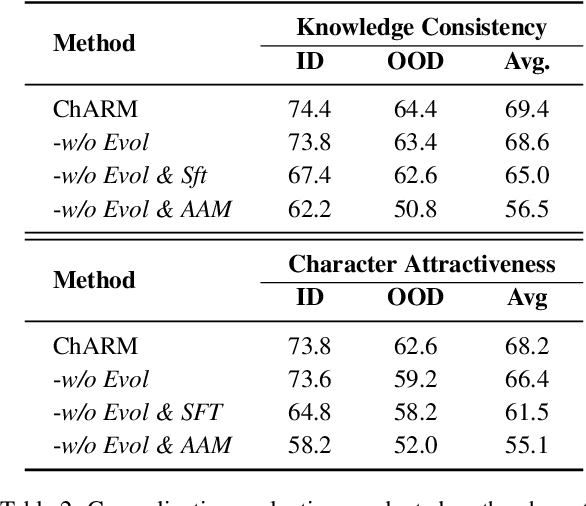
Abstract:Role-Playing Language Agents (RPLAs) aim to simulate characters for realistic and engaging human-computer interactions. However, traditional reward models often struggle with scalability and adapting to subjective conversational preferences. We propose ChARM, a Character-based Act-adaptive Reward Model, addressing these challenges through two innovations: (1) an act-adaptive margin that significantly enhances learning efficiency and generalizability, and (2) a self-evolution mechanism leveraging large-scale unlabeled data to improve training coverage. Additionally, we introduce RoleplayPref, the first large-scale preference dataset specifically for RPLAs, featuring 1,108 characters, 13 subcategories, and 16,888 bilingual dialogues, alongside RoleplayEval, a dedicated evaluation benchmark. Experimental results show a 13% improvement over the conventional Bradley-Terry model in preference rankings. Furthermore, applying ChARM-generated rewards to preference learning techniques (e.g., direct preference optimization) achieves state-of-the-art results on CharacterEval and RoleplayEval. Code and dataset are available at https://github.com/calubkk/ChARM.
EVADE: Multimodal Benchmark for Evasive Content Detection in E-Commerce Applications
May 23, 2025Abstract:E-commerce platforms increasingly rely on Large Language Models (LLMs) and Vision-Language Models (VLMs) to detect illicit or misleading product content. However, these models remain vulnerable to evasive content: inputs (text or images) that superficially comply with platform policies while covertly conveying prohibited claims. Unlike traditional adversarial attacks that induce overt failures, evasive content exploits ambiguity and context, making it far harder to detect. Existing robustness benchmarks provide little guidance for this demanding, real-world challenge. We introduce EVADE, the first expert-curated, Chinese, multimodal benchmark specifically designed to evaluate foundation models on evasive content detection in e-commerce. The dataset contains 2,833 annotated text samples and 13,961 images spanning six demanding product categories, including body shaping, height growth, and health supplements. Two complementary tasks assess distinct capabilities: Single-Violation, which probes fine-grained reasoning under short prompts, and All-in-One, which tests long-context reasoning by merging overlapping policy rules into unified instructions. Notably, the All-in-One setting significantly narrows the performance gap between partial and full-match accuracy, suggesting that clearer rule definitions improve alignment between human and model judgment. We benchmark 26 mainstream LLMs and VLMs and observe substantial performance gaps: even state-of-the-art models frequently misclassify evasive samples. By releasing EVADE and strong baselines, we provide the first rigorous standard for evaluating evasive-content detection, expose fundamental limitations in current multimodal reasoning, and lay the groundwork for safer and more transparent content moderation systems in e-commerce. The dataset is publicly available at https://huggingface.co/datasets/koenshen/EVADE-Bench.
Enhancing Monte Carlo Dropout Performance for Uncertainty Quantification
May 21, 2025Abstract:Knowing the uncertainty associated with the output of a deep neural network is of paramount importance in making trustworthy decisions, particularly in high-stakes fields like medical diagnosis and autonomous systems. Monte Carlo Dropout (MCD) is a widely used method for uncertainty quantification, as it can be easily integrated into various deep architectures. However, conventional MCD often struggles with providing well-calibrated uncertainty estimates. To address this, we introduce innovative frameworks that enhances MCD by integrating different search solutions namely Grey Wolf Optimizer (GWO), Bayesian Optimization (BO), and Particle Swarm Optimization (PSO) as well as an uncertainty-aware loss function, thereby improving the reliability of uncertainty quantification. We conduct comprehensive experiments using different backbones, namely DenseNet121, ResNet50, and VGG16, on various datasets, including Cats vs. Dogs, Myocarditis, Wisconsin, and a synthetic dataset (Circles). Our proposed algorithm outperforms the MCD baseline by 2-3% on average in terms of both conventional accuracy and uncertainty accuracy while achieving significantly better calibration. These results highlight the potential of our approach to enhance the trustworthiness of deep learning models in safety-critical applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge