Zhihao Yang
Training Report of TeleChat3-MoE
Dec 30, 2025Abstract:TeleChat3-MoE is the latest series of TeleChat large language models, featuring a Mixture-of-Experts (MoE) architecture with parameter counts ranging from 105 billion to over one trillion,trained end-to-end on Ascend NPU cluster. This technical report mainly presents the underlying training infrastructure that enables reliable and efficient scaling to frontier model sizes. We detail systematic methodologies for operator-level and end-to-end numerical accuracy verification, ensuring consistency across hardware platforms and distributed parallelism strategies. Furthermore, we introduce a suite of performance optimizations, including interleaved pipeline scheduling, attention-aware data scheduling for long-sequence training,hierarchical and overlapped communication for expert parallelism, and DVM-based operator fusion. A systematic parallelization framework, leveraging analytical estimation and integer linear programming, is also proposed to optimize multi-dimensional parallelism configurations. Additionally, we present methodological approaches to cluster-level optimizations, addressing host- and device-bound bottlenecks during large-scale training tasks. These infrastructure advancements yield significant throughput improvements and near-linear scaling on clusters comprising thousands of devices, providing a robust foundation for large-scale language model development on hardware ecosystems.
Improving the accuracy and generalizability of molecular property regression models with a substructure-substitution-rule-informed framework
Nov 11, 2025Abstract:Artificial Intelligence (AI)-aided drug discovery is an active research field, yet AI models often exhibit poor accuracy in regression tasks for molecular property prediction, and perform catastrophically poorly for out-of-distribution (OOD) molecules. Here, we present MolRuleLoss, a substructure-substitution-rule-informed framework that improves the accuracy and generalizability of multiple molecular property regression models (MPRMs) such as GEM and UniMol for diverse molecular property prediction tasks. MolRuleLoss incorporates partial derivative constraints for substructure substitution rules (SSRs) into an MPRM's loss function. When using GEM models for predicting lipophilicity, water solubility, and solvation-free energy (using lipophilicity, ESOL, and freeSolv datasets from MoleculeNet), the root mean squared error (RMSE) values with and without MolRuleLoss were 0.587 vs. 0.660, 0.777 vs. 0.798, and 1.252 vs. 1.877, respectively, representing 2.6-33.3% performance improvements. We show that both the number and the quality of SSRs contribute to the magnitude of prediction accuracy gains obtained upon adding MolRuleLoss to an MPRM. MolRuleLoss improved the generalizability of MPRMs for "activity cliff" molecules in a lipophilicity prediction task and improved the generalizability of MPRMs for OOD molecules in a melting point prediction task. In a molecular weight prediction task for OOD molecules, MolRuleLoss reduced the RMSE value of a GEM model from 29.507 to 0.007. We also provide a formal demonstration that the upper bound of the variation for property change of SSRs is positively correlated with an MPRM's error. Together, we show that using the MolRuleLoss framework as a bolt-on boosts the prediction accuracy and generalizability of multiple MPRMs, supporting diverse applications in areas like cheminformatics and AI-aided drug discovery.
Modeling the Multivariate Relationship with Contextualized Representations for Effective Human-Object Interaction Detection
Sep 16, 2025Abstract:Human-Object Interaction (HOI) detection aims to simultaneously localize human-object pairs and recognize their interactions. While recent two-stage approaches have made significant progress, they still face challenges due to incomplete context modeling. In this work, we introduce a Contextualized Representation Learning Network that integrates both affordance-guided reasoning and contextual prompts with visual cues to better capture complex interactions. We enhance the conventional HOI detection framework by expanding it beyond simple human-object pairs to include multivariate relationships involving auxiliary entities like tools. Specifically, we explicitly model the functional role (affordance) of these auxiliary objects through triplet structures <human, tool, object>. This enables our model to identify tool-dependent interactions such as 'filling'. Furthermore, the learnable prompt is enriched with instance categories and subsequently integrated with contextual visual features using an attention mechanism. This process aligns language with image content at both global and regional levels. These contextualized representations equip the model with enriched relational cues for more reliable reasoning over complex, context-dependent interactions. Our proposed method demonstrates superior performance on both the HICO-Det and V-COCO datasets in most scenarios. Codes will be released upon acceptance.
Technical Report of TeleChat2, TeleChat2.5 and T1
Jul 24, 2025
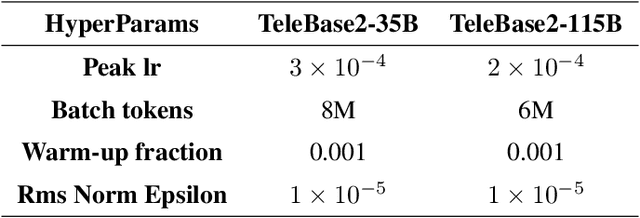
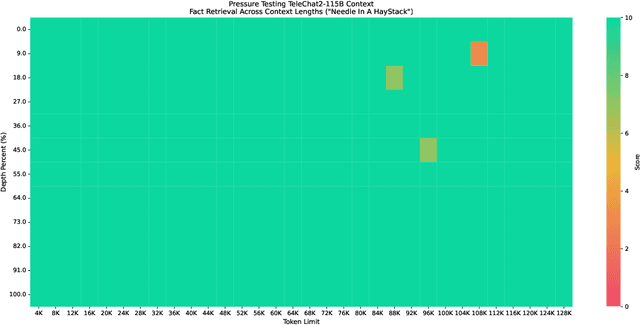
Abstract:We introduce the latest series of TeleChat models: \textbf{TeleChat2}, \textbf{TeleChat2.5}, and \textbf{T1}, offering a significant upgrade over their predecessor, TeleChat. Despite minimal changes to the model architecture, the new series achieves substantial performance gains through enhanced training strategies in both pre-training and post-training stages. The series begins with \textbf{TeleChat2}, which undergoes pretraining on 10 trillion high-quality and diverse tokens. This is followed by Supervised Fine-Tuning (SFT) and Direct Preference Optimization (DPO) to further enhance its capabilities. \textbf{TeleChat2.5} and \textbf{T1} expand the pipeline by incorporating a continual pretraining phase with domain-specific datasets, combined with reinforcement learning (RL) to improve performance in code generation and mathematical reasoning tasks. The \textbf{T1} variant is designed for complex reasoning, supporting long Chain-of-Thought (CoT) reasoning and demonstrating substantial improvements in mathematics and coding. In contrast, \textbf{TeleChat2.5} prioritizes speed, delivering rapid inference. Both flagship models of \textbf{T1} and \textbf{TeleChat2.5} are dense Transformer-based architectures with 115B parameters, showcasing significant advancements in reasoning and general task performance compared to the original TeleChat. Notably, \textbf{T1-115B} outperform proprietary models such as OpenAI's o1-mini and GPT-4o. We publicly release \textbf{TeleChat2}, \textbf{TeleChat2.5} and \textbf{T1}, including post-trained versions with 35B and 115B parameters, to empower developers and researchers with state-of-the-art language models tailored for diverse applications.
EVADE: Multimodal Benchmark for Evasive Content Detection in E-Commerce Applications
May 23, 2025Abstract:E-commerce platforms increasingly rely on Large Language Models (LLMs) and Vision-Language Models (VLMs) to detect illicit or misleading product content. However, these models remain vulnerable to evasive content: inputs (text or images) that superficially comply with platform policies while covertly conveying prohibited claims. Unlike traditional adversarial attacks that induce overt failures, evasive content exploits ambiguity and context, making it far harder to detect. Existing robustness benchmarks provide little guidance for this demanding, real-world challenge. We introduce EVADE, the first expert-curated, Chinese, multimodal benchmark specifically designed to evaluate foundation models on evasive content detection in e-commerce. The dataset contains 2,833 annotated text samples and 13,961 images spanning six demanding product categories, including body shaping, height growth, and health supplements. Two complementary tasks assess distinct capabilities: Single-Violation, which probes fine-grained reasoning under short prompts, and All-in-One, which tests long-context reasoning by merging overlapping policy rules into unified instructions. Notably, the All-in-One setting significantly narrows the performance gap between partial and full-match accuracy, suggesting that clearer rule definitions improve alignment between human and model judgment. We benchmark 26 mainstream LLMs and VLMs and observe substantial performance gaps: even state-of-the-art models frequently misclassify evasive samples. By releasing EVADE and strong baselines, we provide the first rigorous standard for evaluating evasive-content detection, expose fundamental limitations in current multimodal reasoning, and lay the groundwork for safer and more transparent content moderation systems in e-commerce. The dataset is publicly available at https://huggingface.co/datasets/koenshen/EVADE-Bench.
Text-Derived Relational Graph-Enhanced Network for Skeleton-Based Action Segmentation
Mar 19, 2025Abstract:Skeleton-based Temporal Action Segmentation (STAS) aims to segment and recognize various actions from long, untrimmed sequences of human skeletal movements. Current STAS methods typically employ spatio-temporal modeling to establish dependencies among joints as well as frames, and utilize one-hot encoding with cross-entropy loss for frame-wise classification supervision. However, these methods overlook the intrinsic correlations among joints and actions within skeletal features, leading to a limited understanding of human movements. To address this, we propose a Text-Derived Relational Graph-Enhanced Network (TRG-Net) that leverages prior graphs generated by Large Language Models (LLM) to enhance both modeling and supervision. For modeling, the Dynamic Spatio-Temporal Fusion Modeling (DSFM) method incorporates Text-Derived Joint Graphs (TJG) with channel- and frame-level dynamic adaptation to effectively model spatial relations, while integrating spatio-temporal core features during temporal modeling. For supervision, the Absolute-Relative Inter-Class Supervision (ARIS) method employs contrastive learning between action features and text embeddings to regularize the absolute class distributions, and utilizes Text-Derived Action Graphs (TAG) to capture the relative inter-class relationships among action features. Additionally, we propose a Spatial-Aware Enhancement Processing (SAEP) method, which incorporates random joint occlusion and axial rotation to enhance spatial generalization. Performance evaluations on four public datasets demonstrate that TRG-Net achieves state-of-the-art results.
Xpert: Empowering Incident Management with Query Recommendations via Large Language Models
Dec 19, 2023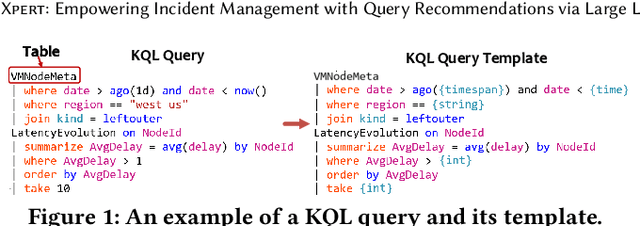

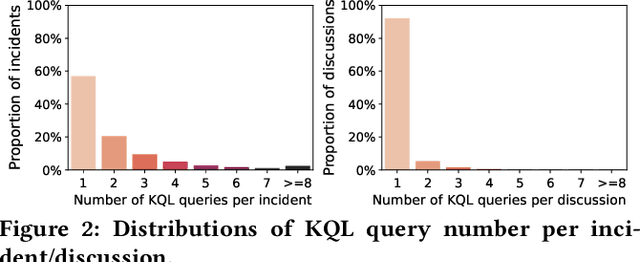

Abstract:Large-scale cloud systems play a pivotal role in modern IT infrastructure. However, incidents occurring within these systems can lead to service disruptions and adversely affect user experience. To swiftly resolve such incidents, on-call engineers depend on crafting domain-specific language (DSL) queries to analyze telemetry data. However, writing these queries can be challenging and time-consuming. This paper presents a thorough empirical study on the utilization of queries of KQL, a DSL employed for incident management in a large-scale cloud management system at Microsoft. The findings obtained underscore the importance and viability of KQL queries recommendation to enhance incident management. Building upon these valuable insights, we introduce Xpert, an end-to-end machine learning framework that automates KQL recommendation process. By leveraging historical incident data and large language models, Xpert generates customized KQL queries tailored to new incidents. Furthermore, Xpert incorporates a novel performance metric called Xcore, enabling a thorough evaluation of query quality from three comprehensive perspectives. We conduct extensive evaluations of Xpert, demonstrating its effectiveness in offline settings. Notably, we deploy Xpert in the real production environment of a large-scale incident management system in Microsoft, validating its efficiency in supporting incident management. To the best of our knowledge, this paper represents the first empirical study of its kind, and Xpert stands as a pioneering DSL query recommendation framework designed for incident management.
Taiyi: A Bilingual Fine-Tuned Large Language Model for Diverse Biomedical Tasks
Nov 20, 2023



Abstract:Recent advancements in large language models (LLMs) have shown promising results across a variety of natural language processing (NLP) tasks. The application of LLMs to specific domains, such as biomedicine, has achieved increased attention. However, most biomedical LLMs focus on enhancing performance in monolingual biomedical question answering and conversation tasks. To further investigate the effectiveness of the LLMs on diverse biomedical NLP tasks in different languages, we present Taiyi, a bilingual (English and Chinese) fine-tuned LLM for diverse biomedical tasks. In this work, we first curated a comprehensive collection of 140 existing biomedical text mining datasets across over 10 task types. Subsequently, a two-stage strategy is proposed for supervised fine-tuning to optimize the model performance across varied tasks. Experimental results on 13 test sets covering named entity recognition, relation extraction, text classification, question answering tasks demonstrate Taiyi achieves superior performance compared to general LLMs. The case study involving additional biomedical NLP tasks further shows Taiyi's considerable potential for bilingual biomedical multi-tasking. The source code, datasets, and model for Taiyi are freely available at https://github.com/DUTIR-BioNLP/Taiyi-LLM.
BokehOrNot: Transforming Bokeh Effect with Image Transformer and Lens Metadata Embedding
Jun 06, 2023



Abstract:Bokeh effect is an optical phenomenon that offers a pleasant visual experience, typically generated by high-end cameras with wide aperture lenses. The task of bokeh effect transformation aims to produce a desired effect in one set of lenses and apertures based on another combination. Current models are limited in their ability to render a specific set of bokeh effects, primarily transformations from sharp to blur. In this paper, we propose a novel universal method for embedding lens metadata into the model and introducing a loss calculation method using alpha masks from the newly released Bokeh Effect Transformation Dataset(BETD) [3]. Based on the above techniques, we propose the BokehOrNot model, which is capable of producing both blur-to-sharp and sharp-to-blur bokeh effect with various combinations of lenses and aperture sizes. Our proposed model outperforms current leading bokeh rendering and image restoration models and renders visually natural bokeh effects. Our code is available at: https://github.com/indicator0/bokehornot.
Biomedical named entity recognition using BERT in the machine reading comprehension framework
Sep 03, 2020Abstract:Recognition of biomedical entities from literature is a challenging research focus, which is the foundation for extracting a large amount of biomedical knowledge existing in unstructured texts into structured formats. Using the sequence labeling framework to implement biomedical named entity recognition (BioNER) is currently a conventional method. This method, however, often cannot take full advantage of the semantic information in the dataset, and the performance is not always satisfactory. In this work, instead of treating the BioNER task as a sequence labeling problem, we formulate it as a machine reading comprehension (MRC) problem. This formulation can introduce more prior knowledge utilizing well-designed queries, and no longer need decoding processes such as conditional random fields (CRF). We conduct experiments on six BioNER datasets, and the experimental results demonstrate the effectiveness of our method. Our method achieves state-of-the-art (SOTA) performance on the BC4CHEMD, BC5CDR-Chem, BC5CDR-Disease, NCBI Disease, BC2GM and JNLPBA datasets, with F1-scores of 92.38%, 94.19%, 87.36%, 90.04%, 84.98% and 78.93%, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge