Saadat Anwar
Developing and Using Special-Purpose Lexicons for Cohort Selection from Clinical Notes
Feb 26, 2019
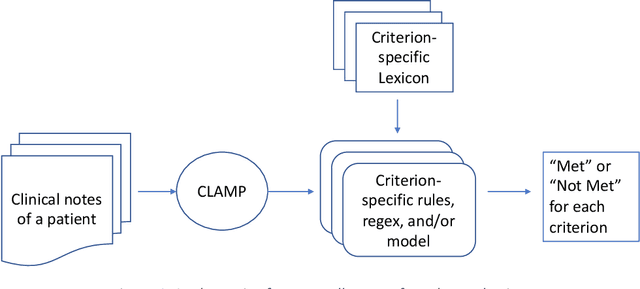

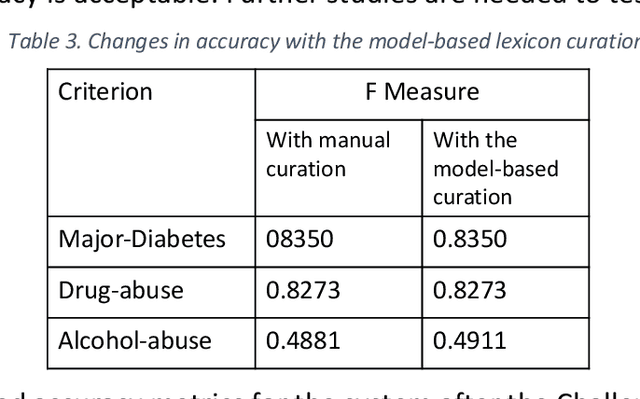
Abstract:Background and Significance: Selecting cohorts for a clinical trial typically requires costly and time-consuming manual chart reviews resulting in poor participation. To help automate the process, National NLP Clinical Challenges (N2C2) conducted a shared challenge by defining 13 criteria for clinical trial cohort selection and by providing training and test datasets. This research was motivated by the N2C2 challenge. Methods: We broke down the task into 13 independent subtasks corresponding to each criterion and implemented subtasks using rules or a supervised machine learning model. Each task critically depended on knowledge resources in the form of task-specific lexicons, for which we developed a novel model-driven approach. The approach allowed us to first expand the lexicon from a seed set and then remove noise from the list, thus improving the accuracy. Results: Our system achieved an overall F measure of 0.9003 at the challenge, and was statistically tied for the first place out of 45 participants. The model-driven lexicon development and further debugging the rules/code on the training set improved overall F measure to 0.9140, overtaking the best numerical result at the challenge. Discussion: Cohort selection, like phenotype extraction and classification, is amenable to rule-based or simple machine learning methods, however, the lexicons involved, such as medication names or medical terms referring to a medical problem, critically determine the overall accuracy. Automated lexicon development has the potential for scalability and accuracy.
Representing, reasoning and answering questions about biological pathways - various applications
Mar 03, 2014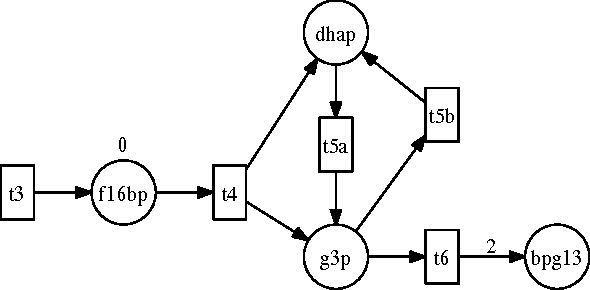
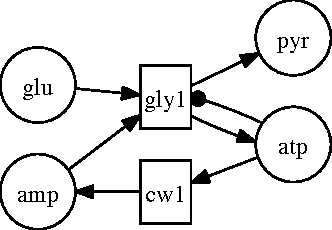
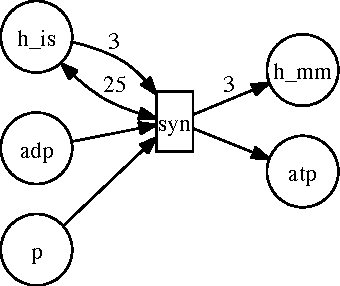
Abstract:Biological organisms are composed of numerous interconnected biochemical processes. Diseases occur when normal functionality of these processes is disrupted. Thus, understanding these biochemical processes and their interrelationships is a primary task in biomedical research and a prerequisite for diagnosing diseases, and drug development. Scientists studying these processes have identified various pathways responsible for drug metabolism, and signal transduction, etc. Newer techniques and speed improvements have resulted in deeper knowledge about these pathways, resulting in refined models that tend to be large and complex, making it difficult for a person to remember all aspects of it. Thus, computer models are needed to analyze them. We want to build such a system that allows modeling of biological systems and pathways in such a way that we can answer questions about them. Many existing models focus on structural and/or factoid questions, using surface-level knowledge that does not require understanding the underlying model. We believe these are not the kind of questions that a biologist may ask someone to test their understanding of the biological processes. We want our system to answer the kind of questions a biologist may ask. Such questions appear in early college level text books. Thus the main goal of our thesis is to develop a system that allows us to encode knowledge about biological pathways and answer such questions about them demonstrating understanding of the pathway. To that end, we develop a language that will allow posing such questions and illustrate the utility of our framework with various applications in the biological domain. We use some existing tools with modifications to accomplish our goal. Finally, we apply our system to real world applications by extracting pathway knowledge from text and answering questions related to drug development.
Encoding Higher Level Extensions of Petri Nets in Answer Set Programming
Jun 24, 2013
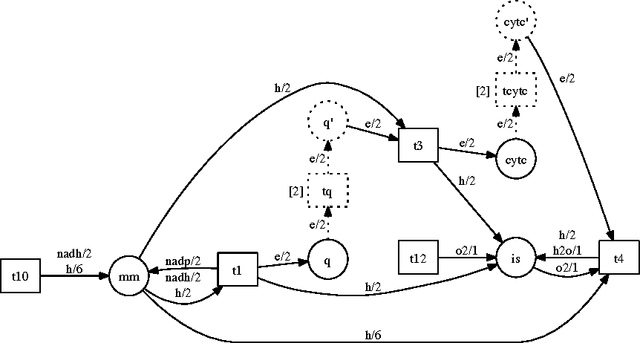
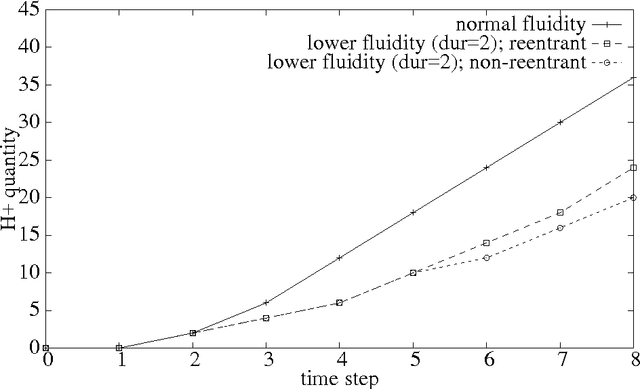
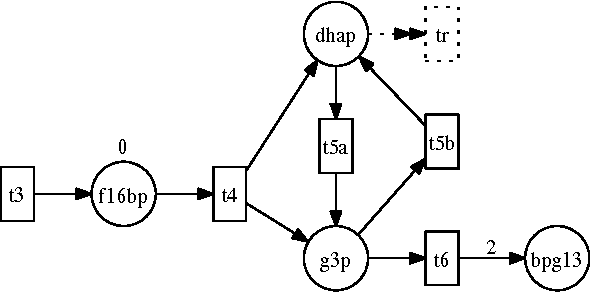
Abstract:Answering realistic questions about biological systems and pathways similar to the ones used by text books to test understanding of students about biological systems is one of our long term research goals. Often these questions require simulation based reasoning. To answer such questions, we need formalisms to build pathway models, add extensions, simulate, and reason with them. We chose Petri Nets and Answer Set Programming (ASP) as suitable formalisms, since Petri Net models are similar to biological pathway diagrams; and ASP provides easy extension and strong reasoning abilities. We found that certain aspects of biological pathways, such as locations and substance types, cannot be represented succinctly using regular Petri Nets. As a result, we need higher level constructs like colored tokens. In this paper, we show how Petri Nets with colored tokens can be encoded in ASP in an intuitive manner, how additional Petri Net extensions can be added by making small code changes, and how this work furthers our long term research goals. Our approach can be adapted to other domains with similar modeling needs.
Encoding Petri Nets in Answer Set Programming for Simulation Based Reasoning
Jun 24, 2013



Abstract:One of our long term research goals is to develop systems to answer realistic questions (e.g., some mentioned in textbooks) about biological pathways that a biologist may ask. To answer such questions we need formalisms that can model pathways, simulate their execution, model intervention to those pathways, and compare simulations under different circumstances. We found Petri Nets to be the starting point of a suitable formalism for the modeling and simulation needs. However, we need to make extensions to the Petri Net model and also reason with multiple simulation runs and parallel state evolutions. Towards that end Answer Set Programming (ASP) implementation of Petri Nets would allow us to do both. In this paper we show how ASP can be used to encode basic Petri Nets in an intuitive manner. We then show how we can modify this encoding to model several Petri Net extensions by making small changes. We then highlight some of the reasoning capabilities that we will use to accomplish our ultimate research goal.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge