Samarth Rawal
Tensor Decomposition of Large-scale Clinical EEGs Reveals Interpretable Patterns of Brain Physiology
Nov 24, 2022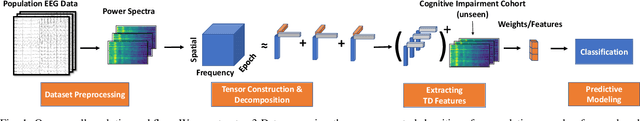
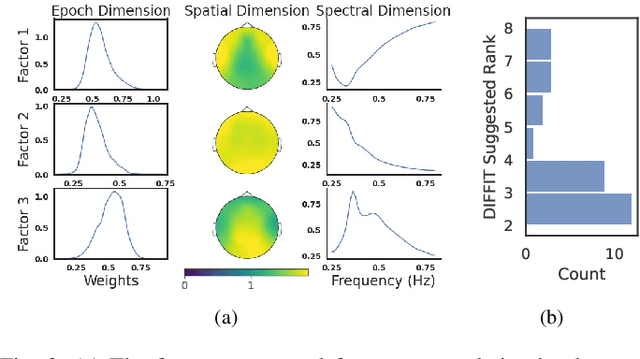
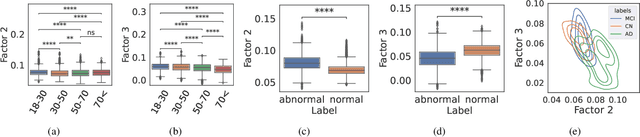
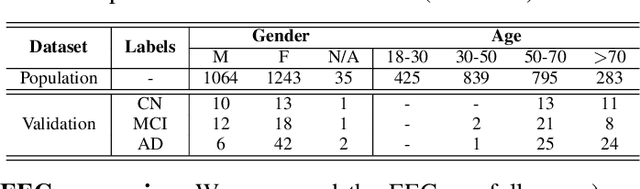
Abstract:Identifying abnormal patterns in electroencephalography (EEG) remains the cornerstone of diagnosing several neurological diseases. The current clinical EEG review process relies heavily on expert visual review, which is unscalable and error-prone. In an effort to augment the expert review process, there is a significant interest in mining population-level EEG patterns using unsupervised approaches. Current approaches rely either on two-dimensional decompositions (e.g., principal and independent component analyses) or deep representation learning (e.g., auto-encoders, self-supervision). However, most approaches do not leverage the natural multi-dimensional structure of EEGs and lack interpretability. In this study, we propose a tensor decomposition approach using the canonical polyadic decomposition to discover a parsimonious set of population-level EEG patterns, retaining the natural multi-dimensional structure of EEGs (time x space x frequency). We then validate their clinical value using a cohort of patients including varying stages of cognitive impairment. Our results show that the discovered patterns reflect physiologically meaningful features and accurately classify the stages of cognitive impairment (healthy vs mild cognitive impairment vs Alzheimer's dementia) with substantially fewer features compared to classical and deep learning-based baselines. We conclude that the decomposition of population-level EEG tensors recovers expert-interpretable EEG patterns that can aid in the study of smaller specialized clinical cohorts.
Assessing Robustness of EEG Representations under Data-shifts via Latent Space and Uncertainty Analysis
Sep 22, 2022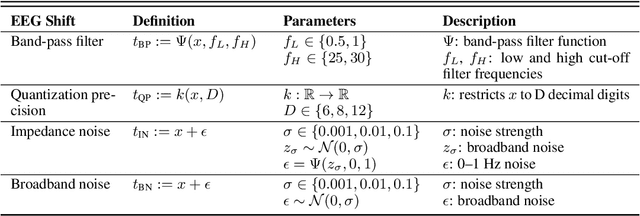
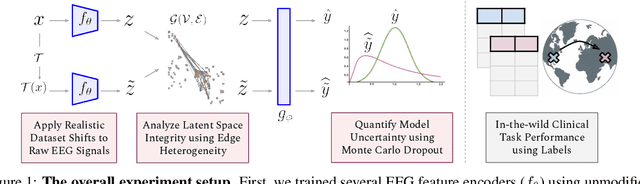
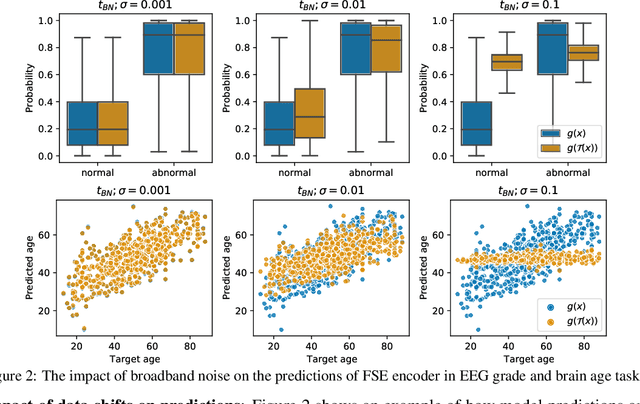
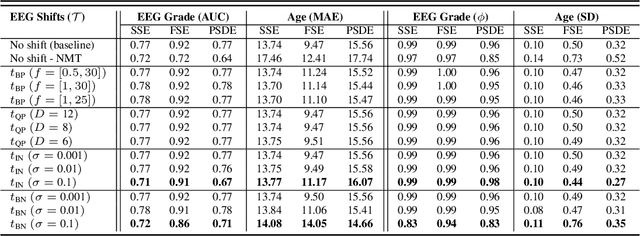
Abstract:The recent availability of large datasets in bio-medicine has inspired the development of representation learning methods for multiple healthcare applications. Despite advances in predictive performance, the clinical utility of such methods is limited when exposed to real-world data. Here we develop model diagnostic measures to detect potential pitfalls during deployment without assuming access to external data. Specifically, we focus on modeling realistic data shifts in electrophysiological signals (EEGs) via data transforms, and extend the conventional task-based evaluations with analyses of a) model's latent space and b) predictive uncertainty, under these transforms. We conduct experiments on multiple EEG feature encoders and two clinically relevant downstream tasks using publicly available large-scale clinical EEGs. Within this experimental setting, our results suggest that measures of latent space integrity and model uncertainty under the proposed data shifts may help anticipate performance degradation during deployment.
SCORE-IT: A Machine Learning-based Tool for Automatic Standardization of EEG Reports
Sep 13, 2021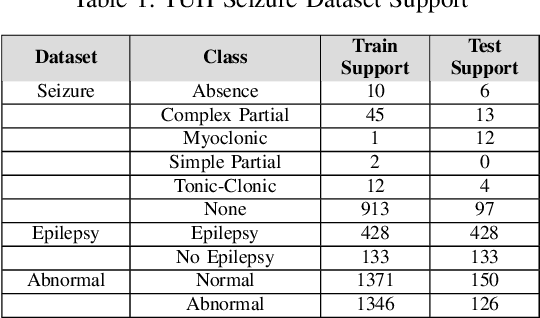

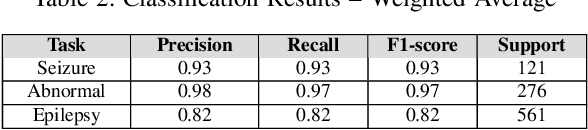
Abstract:Machine learning (ML)-based analysis of electroencephalograms (EEGs) is playing an important role in advancing neurological care. However, the difficulties in automatically extracting useful metadata from clinical records hinder the development of large-scale EEG-based ML models. EEG reports, which are the primary sources of metadata for EEG studies, suffer from lack of standardization. Here we propose a machine learning-based system that automatically extracts components from the SCORE specification from unstructured, natural-language EEG reports. Specifically, our system identifies (1) the type of seizure that was observed in the recording, per physician impression; (2) whether the session recording was normal or abnormal according to physician impression; (3) whether the patient was diagnosed with epilepsy or not. We performed an evaluation of our system using the publicly available TUH EEG corpus and report F1 scores of 0.92, 0.82, and 0.97 for the respective tasks.
Multi-Perspective Semantic Information Retrieval
Sep 03, 2020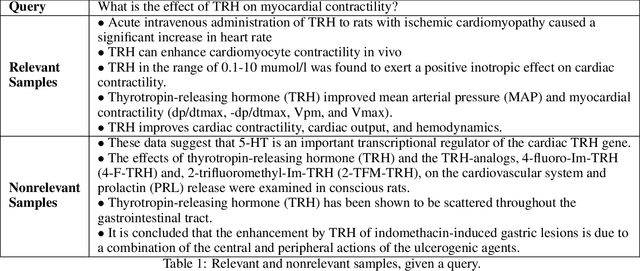

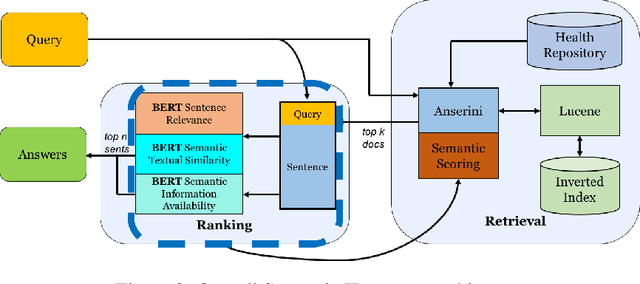

Abstract:Information Retrieval (IR) is the task of obtaining pieces of data (such as documents or snippets of text) that are relevant to a particular query or need from a large repository of information. While a combination of traditional keyword- and modern BERT-based approaches have been shown to be effective in recent work, there are often nuances in identifying what information is "relevant" to a particular query, which can be difficult to properly capture using these systems. This work introduces the concept of a Multi-Perspective IR system, a novel methodology that combines multiple deep learning and traditional IR models to better predict the relevance of a query-sentence pair, along with a standardized framework for tuning this system. This work is evaluated on the BioASQ Biomedical IR + QA challenges.
Multi-Perspective Semantic Information Retrieval in the Biomedical Domain
Jul 17, 2020
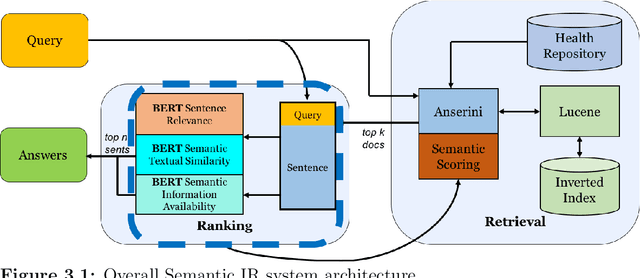
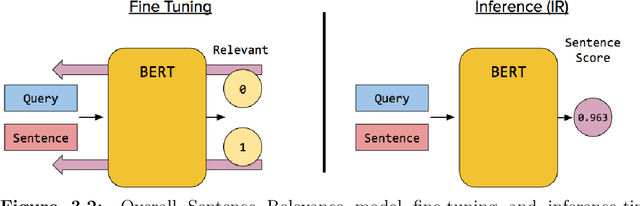
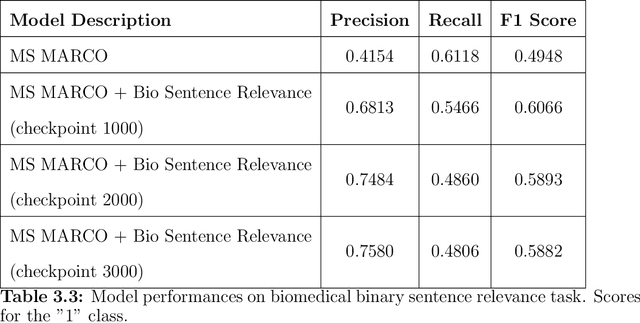
Abstract:Information Retrieval (IR) is the task of obtaining pieces of data (such as documents) that are relevant to a particular query or need from a large repository of information. IR is a valuable component of several downstream Natural Language Processing (NLP) tasks. Practically, IR is at the heart of many widely-used technologies like search engines. While probabilistic ranking functions like the Okapi BM25 function have been utilized in IR systems since the 1970's, modern neural approaches pose certain advantages compared to their classical counterparts. In particular, the release of BERT (Bidirectional Encoder Representations from Transformers) has had a significant impact in the NLP community by demonstrating how the use of a Masked Language Model trained on a large corpus of data can improve a variety of downstream NLP tasks, including sentence classification and passage re-ranking. IR Systems are also important in the biomedical and clinical domains. Given the increasing amount of scientific literature across biomedical domain, the ability find answers to specific clinical queries from a repository of millions of articles is a matter of practical value to medical professionals. Moreover, there are domain-specific challenges present, including handling clinical jargon and evaluating the similarity or relatedness of various medical symptoms when determining the relevance between a query and a sentence. This work presents contributions to several aspects of the Biomedical Semantic Information Retrieval domain. First, it introduces Multi-Perspective Sentence Relevance, a novel methodology of utilizing BERT-based models for contextual IR. The system is evaluated using the BioASQ Biomedical IR Challenge. Finally, practical contributions in the form of a live IR system for medics and a proposed challenge on the Living Systematic Review clinical task are provided.
Developing and Using Special-Purpose Lexicons for Cohort Selection from Clinical Notes
Feb 26, 2019
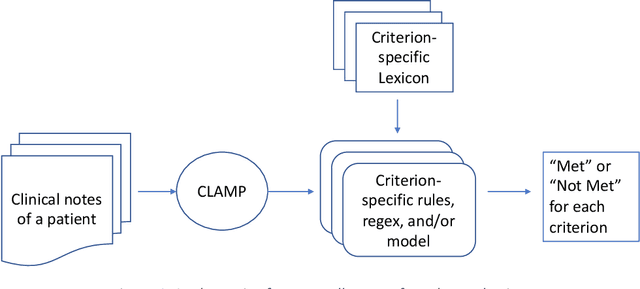

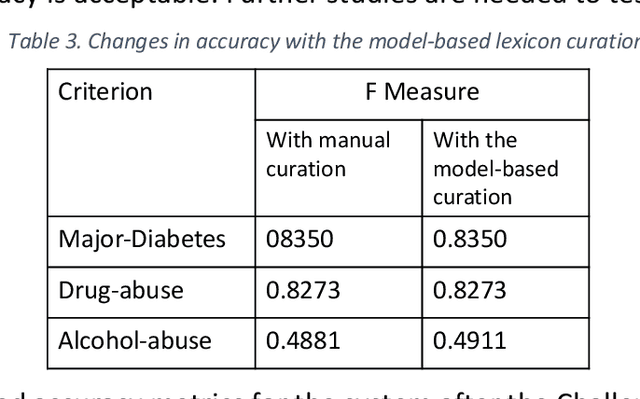
Abstract:Background and Significance: Selecting cohorts for a clinical trial typically requires costly and time-consuming manual chart reviews resulting in poor participation. To help automate the process, National NLP Clinical Challenges (N2C2) conducted a shared challenge by defining 13 criteria for clinical trial cohort selection and by providing training and test datasets. This research was motivated by the N2C2 challenge. Methods: We broke down the task into 13 independent subtasks corresponding to each criterion and implemented subtasks using rules or a supervised machine learning model. Each task critically depended on knowledge resources in the form of task-specific lexicons, for which we developed a novel model-driven approach. The approach allowed us to first expand the lexicon from a seed set and then remove noise from the list, thus improving the accuracy. Results: Our system achieved an overall F measure of 0.9003 at the challenge, and was statistically tied for the first place out of 45 participants. The model-driven lexicon development and further debugging the rules/code on the training set improved overall F measure to 0.9140, overtaking the best numerical result at the challenge. Discussion: Cohort selection, like phenotype extraction and classification, is amenable to rule-based or simple machine learning methods, however, the lexicons involved, such as medication names or medical terms referring to a medical problem, critically determine the overall accuracy. Automated lexicon development has the potential for scalability and accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge