Yogatheesan Varatharajah
Improving Out-of-distribution Human Activity Recognition via IMU-Video Cross-modal Representation Learning
Jul 17, 2025Abstract:Human Activity Recognition (HAR) based on wearable inertial sensors plays a critical role in remote health monitoring. In patients with movement disorders, the ability to detect abnormal patient movements in their home environments can enable continuous optimization of treatments and help alert caretakers as needed. Machine learning approaches have been proposed for HAR tasks using Inertial Measurement Unit (IMU) data; however, most rely on application-specific labels and lack generalizability to data collected in different environments or populations. To address this limitation, we propose a new cross-modal self-supervised pretraining approach to learn representations from large-sale unlabeled IMU-video data and demonstrate improved generalizability in HAR tasks on out of distribution (OOD) IMU datasets, including a dataset collected from patients with Parkinson's disease. Specifically, our results indicate that the proposed cross-modal pretraining approach outperforms the current state-of-the-art IMU-video pretraining approach and IMU-only pretraining under zero-shot and few-shot evaluations. Broadly, our study provides evidence that in highly dynamic data modalities, such as IMU signals, cross-modal pretraining may be a useful tool to learn generalizable data representations. Our software is available at https://github.com/scheshmi/IMU-Video-OOD-HAR.
Foundation Models for Brain Signals: A Critical Review of Current Progress and Future Directions
Jul 15, 2025Abstract:Patterns of electrical brain activity recorded via electroencephalography (EEG) offer immense value for scientific and clinical investigations. The inability of supervised EEG encoders to learn robust EEG patterns and their over-reliance on expensive signal annotations have sparked a transition towards general-purpose self-supervised EEG encoders, i.e., EEG foundation models (EEG-FMs), for robust and scalable EEG feature extraction. However, the real-world readiness of early EEG-FMs and the rubric for long-term research progress remain unclear. A systematic and comprehensive review of first-generation EEG-FMs is therefore necessary to understand the current state-of-the-art and identify key directions for future EEG-FMs. To that end, this study reviews 10 early EEG-FMs and presents a critical synthesis of their methodology, empirical findings, and outstanding research gaps. We find that most EEG-FMs adopt a sequence-based modeling scheme that relies on transformer-based backbones and the reconstruction of masked sequences for self-supervision. However, model evaluations remain heterogeneous and largely limited, making it challenging to assess their practical off-the-shelf utility. In addition to adopting standardized and realistic evaluations, future work should demonstrate more substantial scaling effects and make principled and trustworthy choices throughout the EEG representation learning pipeline. We believe that developing benchmarks, software tools, technical methodologies, and applications in collaboration with domain experts may further advance the translational utility and real-world adoption of EEG-FMs.
Tensor Decomposition of Large-scale Clinical EEGs Reveals Interpretable Patterns of Brain Physiology
Nov 24, 2022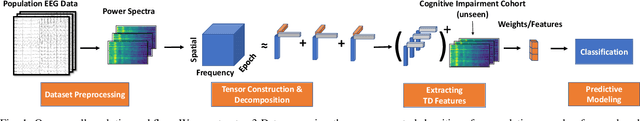
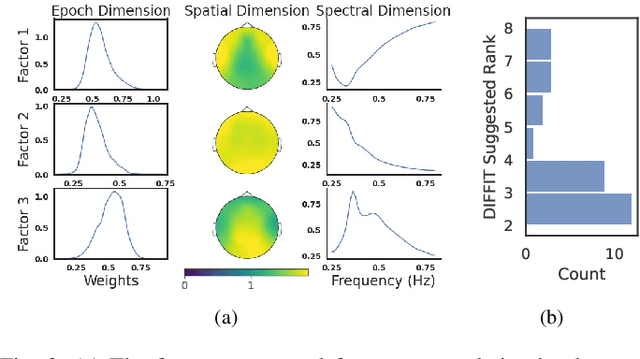
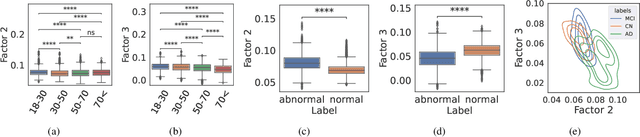
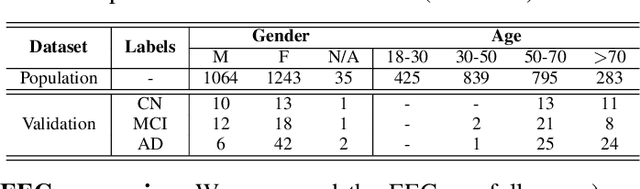
Abstract:Identifying abnormal patterns in electroencephalography (EEG) remains the cornerstone of diagnosing several neurological diseases. The current clinical EEG review process relies heavily on expert visual review, which is unscalable and error-prone. In an effort to augment the expert review process, there is a significant interest in mining population-level EEG patterns using unsupervised approaches. Current approaches rely either on two-dimensional decompositions (e.g., principal and independent component analyses) or deep representation learning (e.g., auto-encoders, self-supervision). However, most approaches do not leverage the natural multi-dimensional structure of EEGs and lack interpretability. In this study, we propose a tensor decomposition approach using the canonical polyadic decomposition to discover a parsimonious set of population-level EEG patterns, retaining the natural multi-dimensional structure of EEGs (time x space x frequency). We then validate their clinical value using a cohort of patients including varying stages of cognitive impairment. Our results show that the discovered patterns reflect physiologically meaningful features and accurately classify the stages of cognitive impairment (healthy vs mild cognitive impairment vs Alzheimer's dementia) with substantially fewer features compared to classical and deep learning-based baselines. We conclude that the decomposition of population-level EEG tensors recovers expert-interpretable EEG patterns that can aid in the study of smaller specialized clinical cohorts.
Assessing Robustness of EEG Representations under Data-shifts via Latent Space and Uncertainty Analysis
Sep 22, 2022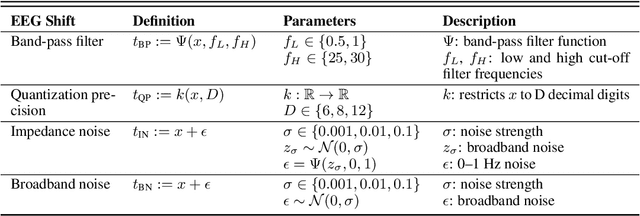
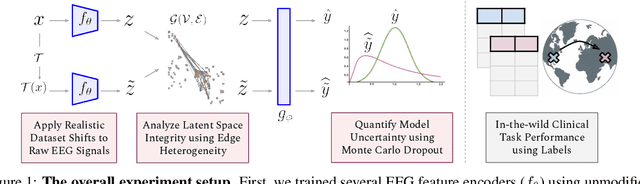
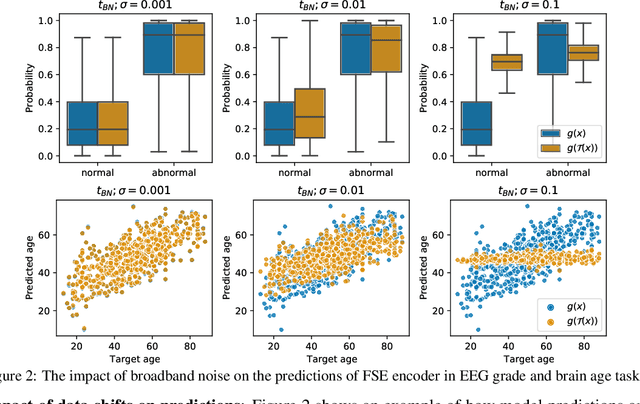
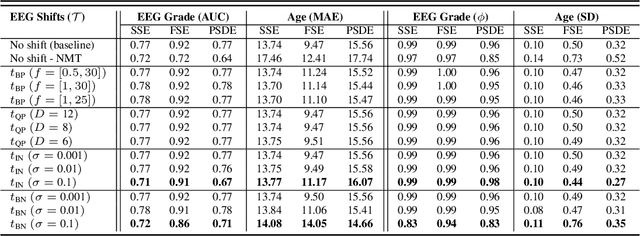
Abstract:The recent availability of large datasets in bio-medicine has inspired the development of representation learning methods for multiple healthcare applications. Despite advances in predictive performance, the clinical utility of such methods is limited when exposed to real-world data. Here we develop model diagnostic measures to detect potential pitfalls during deployment without assuming access to external data. Specifically, we focus on modeling realistic data shifts in electrophysiological signals (EEGs) via data transforms, and extend the conventional task-based evaluations with analyses of a) model's latent space and b) predictive uncertainty, under these transforms. We conduct experiments on multiple EEG feature encoders and two clinically relevant downstream tasks using publicly available large-scale clinical EEGs. Within this experimental setting, our results suggest that measures of latent space integrity and model uncertainty under the proposed data shifts may help anticipate performance degradation during deployment.
SCORE-IT: A Machine Learning-based Tool for Automatic Standardization of EEG Reports
Sep 13, 2021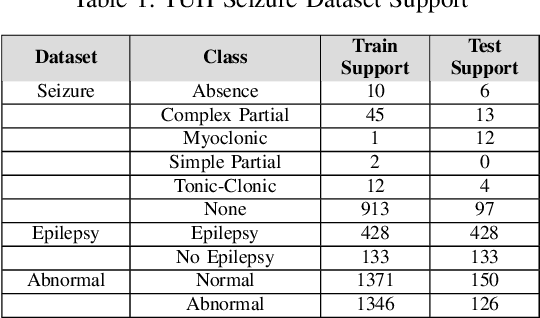

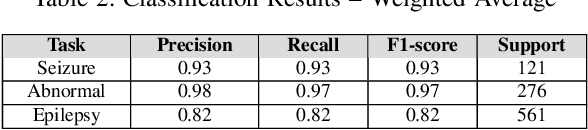
Abstract:Machine learning (ML)-based analysis of electroencephalograms (EEGs) is playing an important role in advancing neurological care. However, the difficulties in automatically extracting useful metadata from clinical records hinder the development of large-scale EEG-based ML models. EEG reports, which are the primary sources of metadata for EEG studies, suffer from lack of standardization. Here we propose a machine learning-based system that automatically extracts components from the SCORE specification from unstructured, natural-language EEG reports. Specifically, our system identifies (1) the type of seizure that was observed in the recording, per physician impression; (2) whether the session recording was normal or abnormal according to physician impression; (3) whether the patient was diagnosed with epilepsy or not. We performed an evaluation of our system using the publicly available TUH EEG corpus and report F1 scores of 0.92, 0.82, and 0.97 for the respective tasks.
EEG-GCNN: Augmenting Electroencephalogram-based Neurological Disease Diagnosis using a Domain-guided Graph Convolutional Neural Network
Nov 17, 2020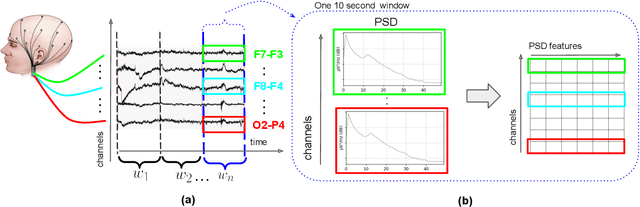

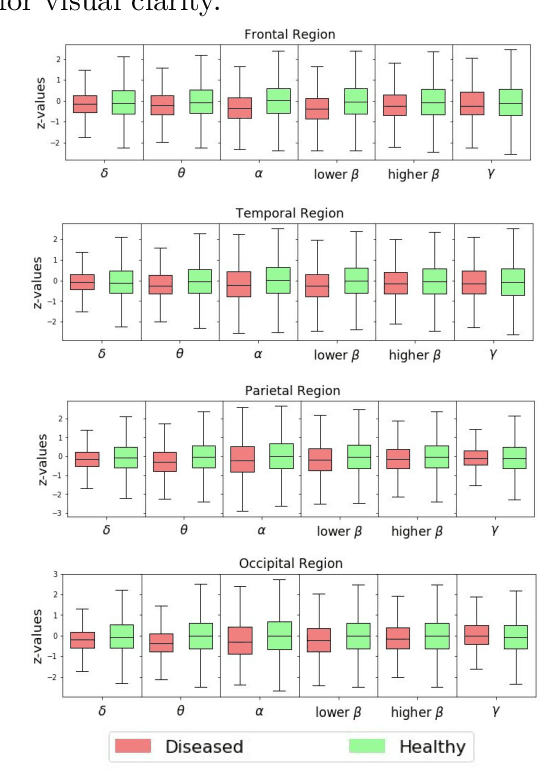

Abstract:This paper presents a novel graph convolutional neural network (GCNN)-based approach for improving the diagnosis of neurological diseases using scalp-electroencephalograms (EEGs). Although EEG is one of the main tests used for neurological-disease diagnosis, the sensitivity of EEG-based expert visual diagnosis remains at $\sim$50\%. This indicates a clear need for advanced methodology to reduce the false negative rate in detecting abnormal scalp-EEGs. In that context, we focus on the problem of distinguishing the abnormal scalp EEGs of patients with neurological diseases, which were originally classified as 'normal' by experts, from the scalp EEGs of healthy individuals. The contributions of this paper are three-fold: 1) we present EEG-GCNN, a novel GCNN model for EEG data that captures both the spatial and functional connectivity between the scalp electrodes, 2) using EEG-GCNN, we perform the first large-scale evaluation of the aforementioned hypothesis, and 3) using two large scalp-EEG databases, we demonstrate that EEG-GCNN significantly outperforms the human baseline and classical machine learning (ML) baselines, with an AUC of 0.90.
Integrating Artificial Intelligence with Real-time Intracranial EEG Monitoring to Automate Interictal Identification of Seizure Onset Zones in Focal Epilepsy
Dec 15, 2018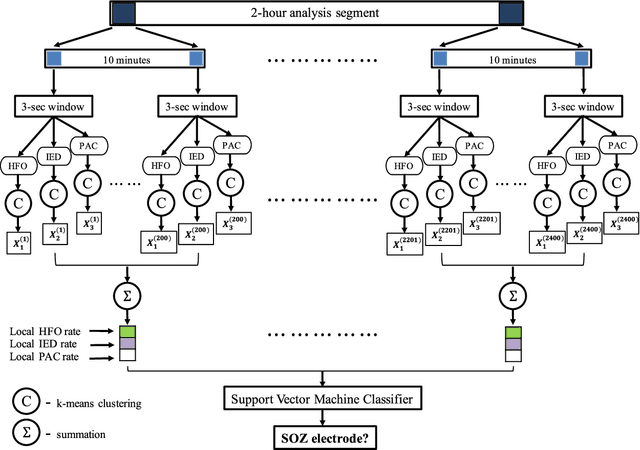

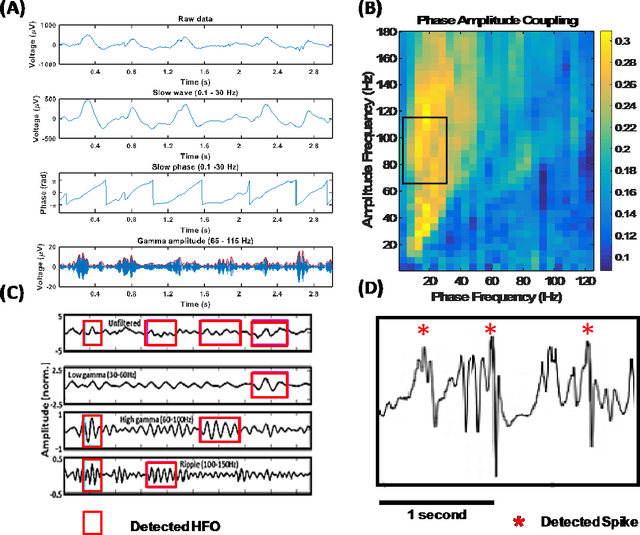
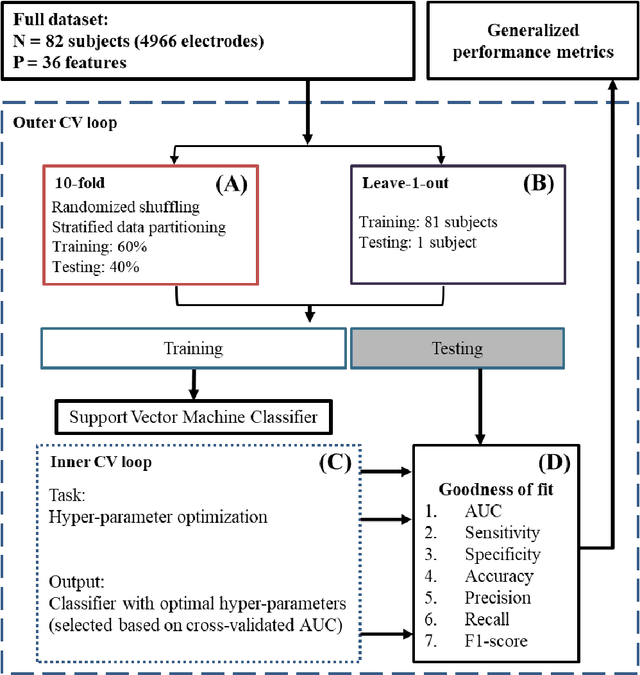
Abstract:An ability to map seizure-generating brain tissue, i.e., the seizure onset zone (SOZ), without recording actual seizures could reduce the duration of invasive EEG monitoring for patients with drug-resistant epilepsy. A widely-adopted practice in the literature is to compare the incidence (events/time) of putative pathological electrophysiological biomarkers associated with epileptic brain tissue with the SOZ determined from spontaneous seizures recorded with intracranial EEG, primarily using a single biomarker. Clinical translation of the previous efforts suffers from their inability to generalize across multiple patients because of (a) the inter-patient variability and (b) the temporal variability in the epileptogenic activity. Here, we report an artificial intelligence-based approach for combining multiple interictal electrophysiological biomarkers and their temporal characteristics as a way of accounting for the above barriers and show that it can reliably identify seizure onset zones in a study cohort of 82 patients who underwent evaluation for drug-resistant epilepsy. Our investigation provides evidence that utilizing the complementary information provided by multiple electrophysiological biomarkers and their temporal characteristics can significantly improve the localization potential compared to previously published single-biomarker incidence-based approaches, resulting in an average area under ROC curve (AUC) value of 0.73 in a cohort of 82 patients. Our results also suggest that recording durations between ninety minutes and two hours are sufficient to localize SOZs with accuracies that may prove clinically relevant. The successful validation of our approach on a large cohort of 82 patients warrants future investigation on the feasibility of utilizing intra-operative EEG monitoring and artificial intelligence to localize epileptogenic brain tissue.
A Contextual-bandit-based Approach for Informed Decision-making in Clinical Trials
Sep 01, 2018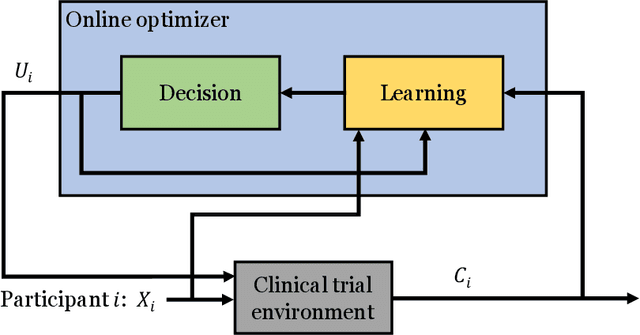
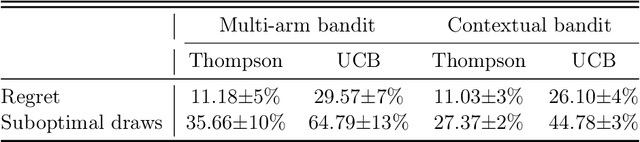
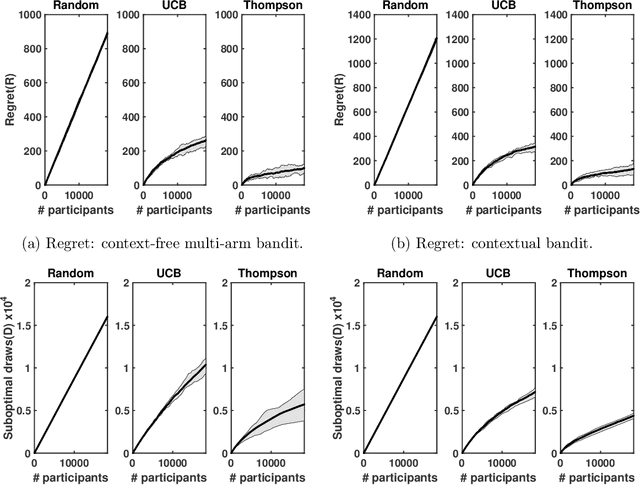
Abstract:Clinical trials involving multiple treatments utilize randomization of the treatment assignments to enable the evaluation of treatment efficacies in an unbiased manner. Such evaluation is performed in post hoc studies that usually use supervised-learning methods that rely on large amounts of data collected in a randomized fashion. That approach often proves to be suboptimal in that some participants may suffer and even die as a result of having not received the most appropriate treatments during the trial. Reinforcement-learning methods improve the situation by making it possible to learn the treatment efficacies dynamically during the course of the trial, and to adapt treatment assignments accordingly. Recent efforts using \textit{multi-arm bandits}, a type of reinforcement-learning methods, have focused on maximizing clinical outcomes for a population that was assumed to be homogeneous. However, those approaches have failed to account for the variability among participants that is becoming increasingly evident as a result of recent clinical-trial-based studies. We present a contextual-bandit-based online treatment optimization algorithm that, in choosing treatments for new participants in the study, takes into account not only the maximization of the clinical outcomes but also the patient characteristics. We evaluated our algorithm using a real clinical trial dataset from the International Stroke Trial. The results of our retrospective analysis indicate that the proposed approach performs significantly better than either a random assignment of treatments (the current gold standard) or a multi-arm-bandit-based approach, providing substantial gains in the percentage of participants who are assigned the most suitable treatments. The contextual-bandit and multi-arm bandit approaches provide 72.63% and 64.34% gains, respectively, compared to a random assignment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge