Brent Berry
Tensor Decomposition of Large-scale Clinical EEGs Reveals Interpretable Patterns of Brain Physiology
Nov 24, 2022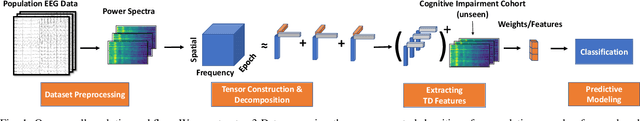
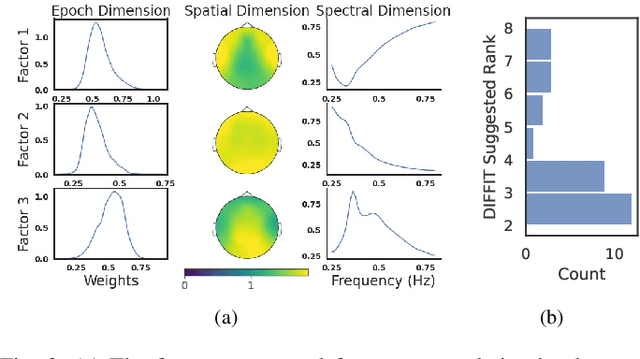
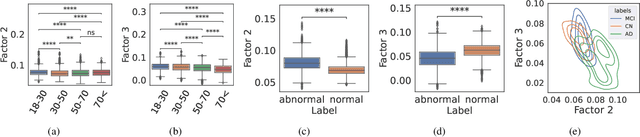
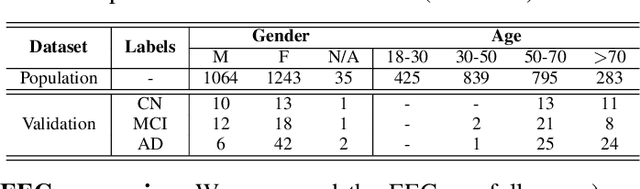
Abstract:Identifying abnormal patterns in electroencephalography (EEG) remains the cornerstone of diagnosing several neurological diseases. The current clinical EEG review process relies heavily on expert visual review, which is unscalable and error-prone. In an effort to augment the expert review process, there is a significant interest in mining population-level EEG patterns using unsupervised approaches. Current approaches rely either on two-dimensional decompositions (e.g., principal and independent component analyses) or deep representation learning (e.g., auto-encoders, self-supervision). However, most approaches do not leverage the natural multi-dimensional structure of EEGs and lack interpretability. In this study, we propose a tensor decomposition approach using the canonical polyadic decomposition to discover a parsimonious set of population-level EEG patterns, retaining the natural multi-dimensional structure of EEGs (time x space x frequency). We then validate their clinical value using a cohort of patients including varying stages of cognitive impairment. Our results show that the discovered patterns reflect physiologically meaningful features and accurately classify the stages of cognitive impairment (healthy vs mild cognitive impairment vs Alzheimer's dementia) with substantially fewer features compared to classical and deep learning-based baselines. We conclude that the decomposition of population-level EEG tensors recovers expert-interpretable EEG patterns that can aid in the study of smaller specialized clinical cohorts.
Integrating Artificial Intelligence with Real-time Intracranial EEG Monitoring to Automate Interictal Identification of Seizure Onset Zones in Focal Epilepsy
Dec 15, 2018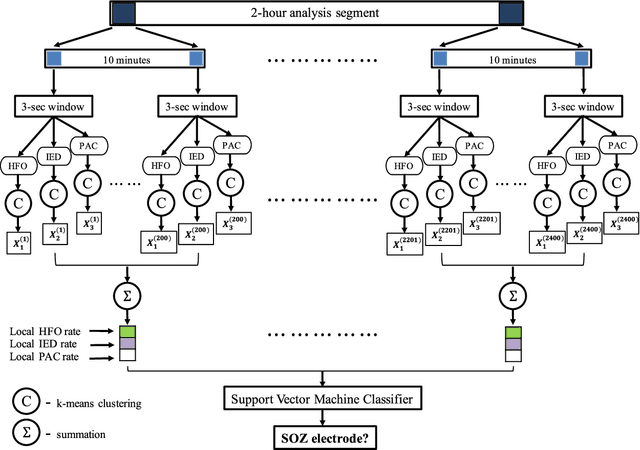

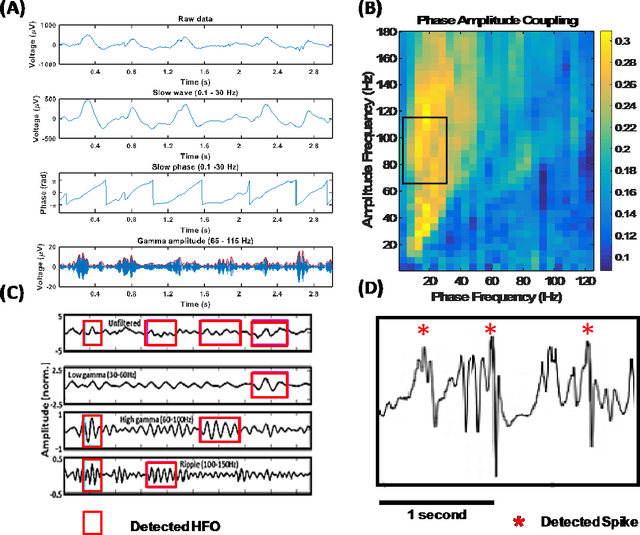
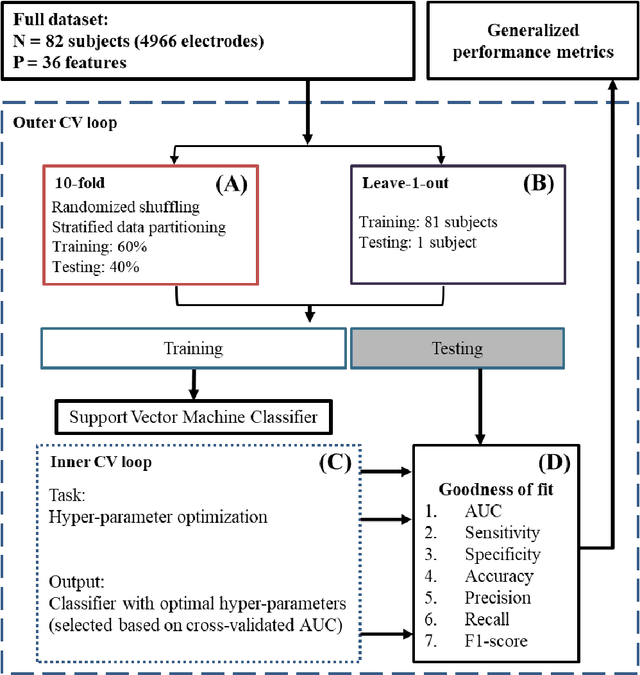
Abstract:An ability to map seizure-generating brain tissue, i.e., the seizure onset zone (SOZ), without recording actual seizures could reduce the duration of invasive EEG monitoring for patients with drug-resistant epilepsy. A widely-adopted practice in the literature is to compare the incidence (events/time) of putative pathological electrophysiological biomarkers associated with epileptic brain tissue with the SOZ determined from spontaneous seizures recorded with intracranial EEG, primarily using a single biomarker. Clinical translation of the previous efforts suffers from their inability to generalize across multiple patients because of (a) the inter-patient variability and (b) the temporal variability in the epileptogenic activity. Here, we report an artificial intelligence-based approach for combining multiple interictal electrophysiological biomarkers and their temporal characteristics as a way of accounting for the above barriers and show that it can reliably identify seizure onset zones in a study cohort of 82 patients who underwent evaluation for drug-resistant epilepsy. Our investigation provides evidence that utilizing the complementary information provided by multiple electrophysiological biomarkers and their temporal characteristics can significantly improve the localization potential compared to previously published single-biomarker incidence-based approaches, resulting in an average area under ROC curve (AUC) value of 0.73 in a cohort of 82 patients. Our results also suggest that recording durations between ninety minutes and two hours are sufficient to localize SOZs with accuracies that may prove clinically relevant. The successful validation of our approach on a large cohort of 82 patients warrants future investigation on the feasibility of utilizing intra-operative EEG monitoring and artificial intelligence to localize epileptogenic brain tissue.
A Contextual-bandit-based Approach for Informed Decision-making in Clinical Trials
Sep 01, 2018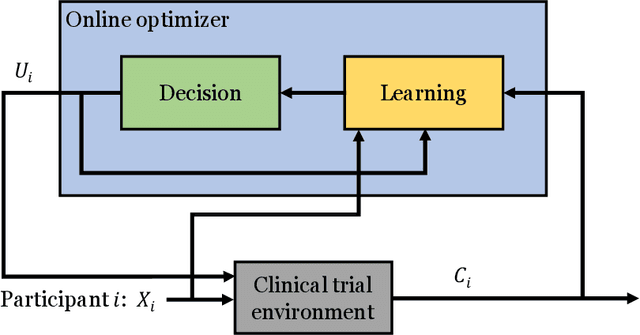
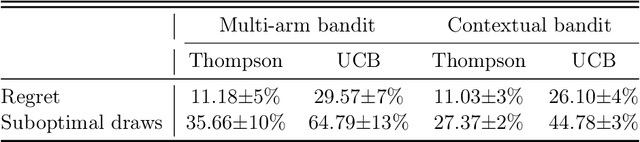
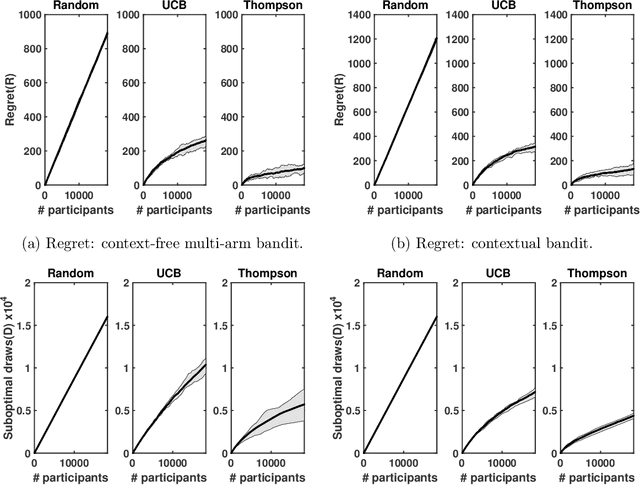
Abstract:Clinical trials involving multiple treatments utilize randomization of the treatment assignments to enable the evaluation of treatment efficacies in an unbiased manner. Such evaluation is performed in post hoc studies that usually use supervised-learning methods that rely on large amounts of data collected in a randomized fashion. That approach often proves to be suboptimal in that some participants may suffer and even die as a result of having not received the most appropriate treatments during the trial. Reinforcement-learning methods improve the situation by making it possible to learn the treatment efficacies dynamically during the course of the trial, and to adapt treatment assignments accordingly. Recent efforts using \textit{multi-arm bandits}, a type of reinforcement-learning methods, have focused on maximizing clinical outcomes for a population that was assumed to be homogeneous. However, those approaches have failed to account for the variability among participants that is becoming increasingly evident as a result of recent clinical-trial-based studies. We present a contextual-bandit-based online treatment optimization algorithm that, in choosing treatments for new participants in the study, takes into account not only the maximization of the clinical outcomes but also the patient characteristics. We evaluated our algorithm using a real clinical trial dataset from the International Stroke Trial. The results of our retrospective analysis indicate that the proposed approach performs significantly better than either a random assignment of treatments (the current gold standard) or a multi-arm-bandit-based approach, providing substantial gains in the percentage of participants who are assigned the most suitable treatments. The contextual-bandit and multi-arm bandit approaches provide 72.63% and 64.34% gains, respectively, compared to a random assignment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge