Alain Jungo
Unsupervised out-of-distribution detection for safer robotically guided retinal microsurgery
May 03, 2023Abstract:Purpose: A fundamental problem in designing safe machine learning systems is identifying when samples presented to a deployed model differ from those observed at training time. Detecting so-called out-of-distribution (OoD) samples is crucial in safety-critical applications such as robotically guided retinal microsurgery, where distances between the instrument and the retina are derived from sequences of 1D images that are acquired by an instrument-integrated optical coherence tomography (iiOCT) probe. Methods: This work investigates the feasibility of using an OoD detector to identify when images from the iiOCT probe are inappropriate for subsequent machine learning-based distance estimation. We show how a simple OoD detector based on the Mahalanobis distance can successfully reject corrupted samples coming from real-world ex vivo porcine eyes. Results: Our results demonstrate that the proposed approach can successfully detect OoD samples and help maintain the performance of the downstream task within reasonable levels. MahaAD outperformed a supervised approach trained on the same kind of corruptions and achieved the best performance in detecting OoD cases from a collection of iiOCT samples with real-world corruptions. Conclusion: The results indicate that detecting corrupted iiOCT data through OoD detection is feasible and does not need prior knowledge of possible corruptions. Consequently, MahaAD could aid in ensuring patient safety during robotically guided microsurgery by preventing deployed prediction models from estimating distances that put the patient at risk.
The MICCAI Hackathon on reproducibility, diversity, and selection of papers at the MICCAI conference
Mar 04, 2021
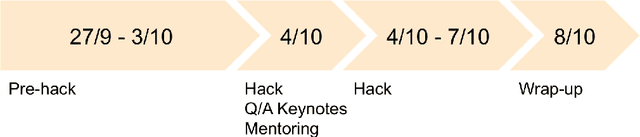
Abstract:The MICCAI conference has encountered tremendous growth over the last years in terms of the size of the community, as well as the number of contributions and their technical success. With this growth, however, come new challenges for the community. Methods are more difficult to reproduce and the ever-increasing number of paper submissions to the MICCAI conference poses new questions regarding the selection process and the diversity of topics. To exchange, discuss, and find novel and creative solutions to these challenges, a new format of a hackathon was initiated as a satellite event at the MICCAI 2020 conference: The MICCAI Hackathon. The first edition of the MICCAI Hackathon covered the topics reproducibility, diversity, and selection of MICCAI papers. In the manner of a small think-tank, participants collaborated to find solutions to these challenges. In this report, we summarize the insights from the MICCAI Hackathon into immediate and long-term measures to address these challenges. The proposed measures can be seen as starting points and guidelines for discussions and actions to possibly improve the MICCAI conference with regards to reproducibility, diversity, and selection of papers.
pymia: A Python package for data handling and evaluation in deep learning-based medical image analysis
Oct 07, 2020



Abstract:Background and Objective: Deep learning enables tremendous progress in medical image analysis. One driving force of this progress are open-source frameworks like TensorFlow and PyTorch. However, these frameworks rarely address issues specific to the domain of medical image analysis, such as 3-D data handling and distance metrics for evaluation. pymia, an open-source Python package, tries to address these issues by providing flexible data handling and evaluation independent of the deep learning framework. Methods: The pymia package provides data handling and evaluation functionalities. The data handling allows flexible medical image handling in every commonly used format (e.g., 2-D, 2.5-D, and 3-D; full- or patch-wise). Even data beyond images like demographics or clinical reports can easily be integrated into deep learning pipelines. The evaluation allows stand-alone result calculation and reporting, as well as performance monitoring during training using a vast amount of domain-specific metrics for segmentation, reconstruction, and regression. Results: The pymia package is highly flexible, allows for fast prototyping, and reduces the burden of implementing data handling routines and evaluation methods. While data handling and evaluation are independent of the deep learning framework used, they can easily be integrated into TensorFlow and PyTorch pipelines. The developed package was successfully used in a variety of research projects for segmentation, reconstruction, and regression. Conclusions: The pymia package fills the gap of current deep learning frameworks regarding data handling and evaluation in medical image analysis. It is available at https://github.com/rundherum/pymia and can directly be installed from the Python Package Index using pip install pymia.
Learning Bloch Simulations for MR Fingerprinting by Invertible Neural Networks
Aug 10, 2020



Abstract:Magnetic resonance fingerprinting (MRF) enables fast and multiparametric MR imaging. Despite fast acquisition, the state-of-the-art reconstruction of MRF based on dictionary matching is slow and lacks scalability. To overcome these limitations, neural network (NN) approaches estimating MR parameters from fingerprints have been proposed recently. Here, we revisit NN-based MRF reconstruction to jointly learn the forward process from MR parameters to fingerprints and the backward process from fingerprints to MR parameters by leveraging invertible neural networks (INNs). As a proof-of-concept, we perform various experiments showing the benefit of learning the forward process, i.e., the Bloch simulations, for improved MR parameter estimation. The benefit especially accentuates when MR parameter estimation is difficult due to MR physical restrictions. Therefore, INNs might be a feasible alternative to the current solely backward-based NNs for MRF reconstruction.
Spatially Regularized Parametric Map Reconstruction for Fast Magnetic Resonance Fingerprinting
Nov 09, 2019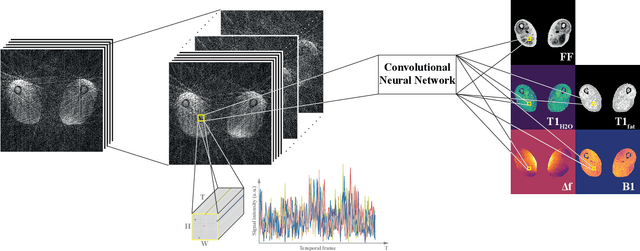

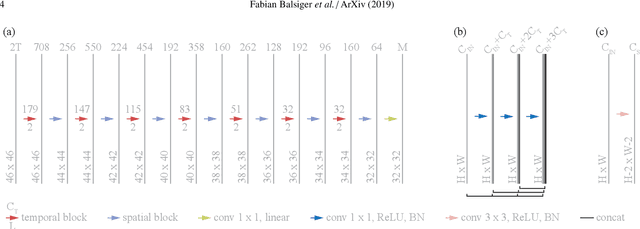

Abstract:Magnetic resonance fingerprinting (MRF) provides a unique concept for simultaneous and fast acquisition of multiple quantitative MR parameters. Despite acquisition efficiency, adoption of MRF into the clinics is hindered by its dictionary-based reconstruction, which is computationally demanding and lacks scalability. Here, we propose a convolutional neural network-based reconstruction, which enables both accurate and fast reconstruction of parametric maps, and is adaptable based on the needs of spatial regularization and the capacity for the reconstruction. We evaluated the method using MRF T1-FF, an MRF sequence for T1 relaxation time of water and fat fraction mapping. We demonstrate the method's performance on a highly heterogeneous dataset consisting of 164 patients with various neuromuscular diseases imaged at thighs and legs. We empirically show the benefit of incorporating spatial regularization during the reconstruction and demonstrate that the method learns meaningful features from MR physics perspective. Further, we investigate the ability of the method to handle highly heterogeneous morphometric variations and its generalization to anatomical regions unseen during training. The obtained results outperform the state-of-the-art in deep learning-based MRF reconstruction. Coupled with fast MRF sequences, the proposed method has the potential of enabling multiparametric MR imaging in clinically feasible time.
Assessing Reliability and Challenges of Uncertainty Estimations for Medical Image Segmentation
Jul 07, 2019

Abstract:Despite the recent improvements in overall accuracy, deep learning systems still exhibit low levels of robustness. Detecting possible failures is critical for a successful clinical integration of these systems, where each data point corresponds to an individual patient. Uncertainty measures are a promising direction to improve failure detection since they provide a measure of a system's confidence. Although many uncertainty estimation methods have been proposed for deep learning, little is known on their benefits and current challenges for medical image segmentation. Therefore, we report results of evaluating common voxel-wise uncertainty measures with respect to their reliability, and limitations on two medical image segmentation datasets. Results show that current uncertainty methods perform similarly and although they are well-calibrated at the dataset level, they tend to be miscalibrated at subject-level. Therefore, the reliability of uncertainty estimates is compromised, highlighting the importance of developing subject-wise uncertainty estimations. Additionally, among the benchmarked methods, we found auxiliary networks to be a valid alternative to common uncertainty methods since they can be applied to any previously trained segmentation model.
Deep Learning versus Classical Regression for Brain Tumor Patient Survival Prediction
Nov 12, 2018



Abstract:Deep learning for regression tasks on medical imaging data has shown promising results. However, compared to other approaches, their power is strongly linked to the dataset size. In this study, we evaluate 3D-convolutional neural networks (CNNs) and classical regression methods with hand-crafted features for survival time regression of patients with high grade brain tumors. The tested CNNs for regression showed promising but unstable results. The best performing deep learning approach reached an accuracy of 51.5% on held-out samples of the training set. All tested deep learning experiments were outperformed by a Support Vector Classifier (SVC) using 30 radiomic features. The investigated features included intensity, shape, location and deep features. The submitted method to the BraTS 2018 survival prediction challenge is an ensemble of SVCs, which reached a cross-validated accuracy of 72.2% on the BraTS 2018 training set, 57.1% on the validation set, and 42.9% on the testing set. The results suggest that more training data is necessary for a stable performance of a CNN model for direct regression from magnetic resonance images, and that non-imaging clinical patient information is crucial along with imaging information.
Uncertainty-driven Sanity Check: Application to Postoperative Brain Tumor Cavity Segmentation
Jun 08, 2018



Abstract:Uncertainty estimates of modern neuronal networks provide additional information next to the computed predictions and are thus expected to improve the understanding of the underlying model. Reliable uncertainties are particularly interesting for safety-critical computer-assisted applications in medicine, e.g., neurosurgical interventions and radiotherapy planning. We propose an uncertainty-driven sanity check for the identification of segmentation results that need particular expert review. Our method uses a fully-convolutional neural network and computes uncertainty estimates by the principle of Monte Carlo dropout. We evaluate the performance of the proposed method on a clinical dataset with 30 postoperative brain tumor images. The method can segment the highly inhomogeneous resection cavities accurately (Dice coefficients 0.792 $\pm$ 0.154). Furthermore, the proposed sanity check is able to detect the worst segmentation and three out of the four outliers. The results highlight the potential of using the additional information from the model's parameter uncertainty to validate the segmentation performance of a deep learning model.
On the Effect of Inter-observer Variability for a Reliable Estimation of Uncertainty of Medical Image Segmentation
Jun 07, 2018



Abstract:Uncertainty estimation methods are expected to improve the understanding and quality of computer-assisted methods used in medical applications (e.g., neurosurgical interventions, radiotherapy planning), where automated medical image segmentation is crucial. In supervised machine learning, a common practice to generate ground truth label data is to merge observer annotations. However, as many medical image tasks show a high inter-observer variability resulting from factors such as image quality, different levels of user expertise and domain knowledge, little is known as to how inter-observer variability and commonly used fusion methods affect the estimation of uncertainty of automated image segmentation. In this paper we analyze the effect of common image label fusion techniques on uncertainty estimation, and propose to learn the uncertainty among observers. The results highlight the negative effect of fusion methods applied in deep learning, to obtain reliable estimates of segmentation uncertainty. Additionally, we show that the learned observers' uncertainty can be combined with current standard Monte Carlo dropout Bayesian neural networks to characterize uncertainty of model's parameters.
Perturb-and-MPM: Quantifying Segmentation Uncertainty in Dense Multi-Label CRFs
Mar 02, 2017



Abstract:This paper proposes a novel approach for uncertainty quantification in dense Conditional Random Fields (CRFs). The presented approach, called Perturb-and-MPM, enables efficient, approximate sampling from dense multi-label CRFs via random perturbations. An analytic error analysis was performed which identified the main cause of approximation error as well as showed that the error is bounded. Spatial uncertainty maps can be derived from the Perturb-and-MPM model, which can be used to visualize uncertainty in image segmentation results. The method is validated on synthetic and clinical Magnetic Resonance Imaging data. The effectiveness of the approach is demonstrated on the challenging problem of segmenting the tumor core in glioblastoma. We found that areas of high uncertainty correspond well to wrongly segmented image regions. Furthermore, we demonstrate the potential use of uncertainty maps to refine imaging biomarkers in the case of extent of resection and residual tumor volume in brain tumor patients.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge