Pierre G. Carlier
UPMC
Spatially Regularized Parametric Map Reconstruction for Fast Magnetic Resonance Fingerprinting
Nov 09, 2019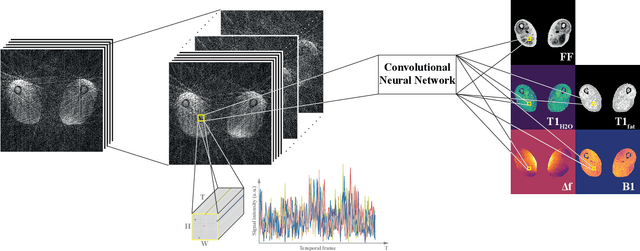

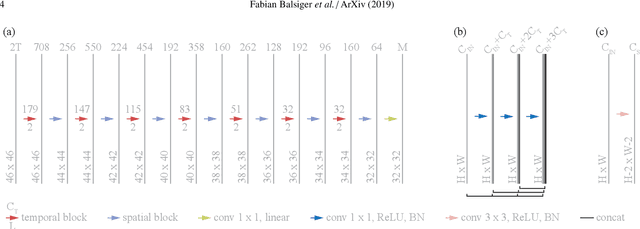

Abstract:Magnetic resonance fingerprinting (MRF) provides a unique concept for simultaneous and fast acquisition of multiple quantitative MR parameters. Despite acquisition efficiency, adoption of MRF into the clinics is hindered by its dictionary-based reconstruction, which is computationally demanding and lacks scalability. Here, we propose a convolutional neural network-based reconstruction, which enables both accurate and fast reconstruction of parametric maps, and is adaptable based on the needs of spatial regularization and the capacity for the reconstruction. We evaluated the method using MRF T1-FF, an MRF sequence for T1 relaxation time of water and fat fraction mapping. We demonstrate the method's performance on a highly heterogeneous dataset consisting of 164 patients with various neuromuscular diseases imaged at thighs and legs. We empirically show the benefit of incorporating spatial regularization during the reconstruction and demonstrate that the method learns meaningful features from MR physics perspective. Further, we investigate the ability of the method to handle highly heterogeneous morphometric variations and its generalization to anatomical regions unseen during training. The obtained results outperform the state-of-the-art in deep learning-based MRF reconstruction. Coupled with fast MRF sequences, the proposed method has the potential of enabling multiparametric MR imaging in clinically feasible time.
Discriminative Parameter Estimation for Random Walks Segmentation
Aug 30, 2013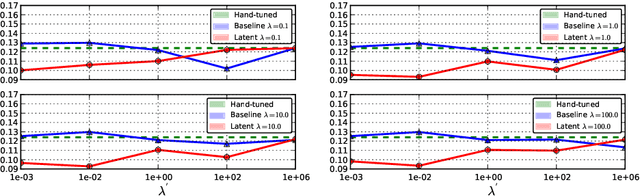
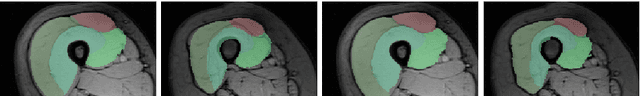
Abstract:The Random Walks (RW) algorithm is one of the most e - cient and easy-to-use probabilistic segmentation methods. By combining contrast terms with prior terms, it provides accurate segmentations of medical images in a fully automated manner. However, one of the main drawbacks of using the RW algorithm is that its parameters have to be hand-tuned. we propose a novel discriminative learning framework that estimates the parameters using a training dataset. The main challenge we face is that the training samples are not fully supervised. Speci cally, they provide a hard segmentation of the images, instead of a proba- bilistic segmentation. We overcome this challenge by treating the opti- mal probabilistic segmentation that is compatible with the given hard segmentation as a latent variable. This allows us to employ the latent support vector machine formulation for parameter estimation. We show that our approach signi cantly outperforms the baseline methods on a challenging dataset consisting of real clinical 3D MRI volumes of skeletal muscles.
Discriminative Parameter Estimation for Random Walks Segmentation: Technical Report
Jun 05, 2013

Abstract:The Random Walks (RW) algorithm is one of the most e - cient and easy-to-use probabilistic segmentation methods. By combining contrast terms with prior terms, it provides accurate segmentations of medical images in a fully automated manner. However, one of the main drawbacks of using the RW algorithm is that its parameters have to be hand-tuned. we propose a novel discriminative learning framework that estimates the parameters using a training dataset. The main challenge we face is that the training samples are not fully supervised. Speci cally, they provide a hard segmentation of the images, instead of a proba-bilistic segmentation. We overcome this challenge by treating the optimal probabilistic segmentation that is compatible with the given hard segmentation as a latent variable. This allows us to employ the latent support vector machine formulation for parameter estimation. We show that our approach signi cantly outperforms the baseline methods on a challenging dataset consisting of real clinical 3D MRI volumes of skeletal muscles.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge