Pierre-Yves Baudin
INRIA Saclay - Ile de France
Upper-body free-breathing Magnetic Resonance Fingerprinting applied to the quantification of water T1 and fat fraction
Sep 24, 2024Abstract:Over the past decade, Magnetic Resonance Fingerprinting (MRF) has emerged as an efficient paradigm for the rapid and simultaneous quantification of multiple MRI parameters, including fat fraction (FF), water T1 ($T1_{H2O}$), water T2 ($T2_{H2O}$), and fat T1 ($T1_{fat}$). These parameters serve as promising imaging biomarkers in various anatomical targets such as the heart, liver, and skeletal muscles. However, measuring these parameters in the upper body poses challenges due to physiological motion, particularly respiratory motion. In this work, we propose a novel approach, motion-corrected (MoCo) MRF T1-FF, which estimates the motion field using an optimized preliminary motion scan and uses it to correct the MRF acquisition data before dictionary search for reconstructing motion-corrected FF and $T1_{H2O}$ parametric maps of the upper-body region. We validated this framework using an $\textit{in vivo}$ dataset comprising ten healthy volunteers and a 10-year-old boy with Duchenne muscular dystrophy. At the ROI level, in regions minimally affected by motion, no significant bias was observed between the uncorrected and MoCo reconstructions for FF (mean difference of -0.7%) and $T1_{H2O}$ (-4.9 ms) values. Moreover, MoCo MRF T1-FF significantly reduced the standard deviations of distributions assessed in these regions, indicating improved precision. Notably, in regions heavily affected by motion, such as respiratory muscles, liver, and kidneys, the MRF parametric maps exhibited a marked reduction in motion blurring and streaking artifacts after motion correction. Furthermore, the diaphragm was consistently discernible on parametric maps after motion correction. This approach lays the groundwork for the joint 3D quantification of FF and $T1_{H2O}$ in regions that are rarely studied, such as the respiratory muscles, particularly the intercostal muscles and diaphragm.
Discriminative Parameter Estimation for Random Walks Segmentation
Aug 30, 2013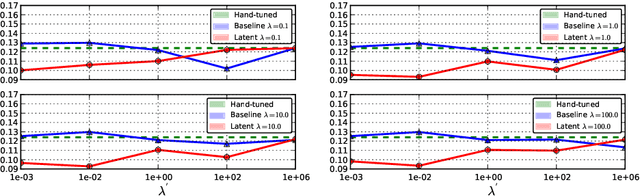
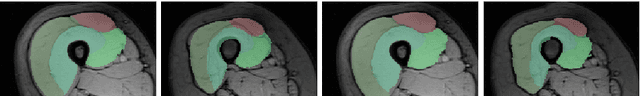
Abstract:The Random Walks (RW) algorithm is one of the most e - cient and easy-to-use probabilistic segmentation methods. By combining contrast terms with prior terms, it provides accurate segmentations of medical images in a fully automated manner. However, one of the main drawbacks of using the RW algorithm is that its parameters have to be hand-tuned. we propose a novel discriminative learning framework that estimates the parameters using a training dataset. The main challenge we face is that the training samples are not fully supervised. Speci cally, they provide a hard segmentation of the images, instead of a proba- bilistic segmentation. We overcome this challenge by treating the opti- mal probabilistic segmentation that is compatible with the given hard segmentation as a latent variable. This allows us to employ the latent support vector machine formulation for parameter estimation. We show that our approach signi cantly outperforms the baseline methods on a challenging dataset consisting of real clinical 3D MRI volumes of skeletal muscles.
Discriminative Parameter Estimation for Random Walks Segmentation: Technical Report
Jun 05, 2013

Abstract:The Random Walks (RW) algorithm is one of the most e - cient and easy-to-use probabilistic segmentation methods. By combining contrast terms with prior terms, it provides accurate segmentations of medical images in a fully automated manner. However, one of the main drawbacks of using the RW algorithm is that its parameters have to be hand-tuned. we propose a novel discriminative learning framework that estimates the parameters using a training dataset. The main challenge we face is that the training samples are not fully supervised. Speci cally, they provide a hard segmentation of the images, instead of a proba-bilistic segmentation. We overcome this challenge by treating the optimal probabilistic segmentation that is compatible with the given hard segmentation as a latent variable. This allows us to employ the latent support vector machine formulation for parameter estimation. We show that our approach signi cantly outperforms the baseline methods on a challenging dataset consisting of real clinical 3D MRI volumes of skeletal muscles.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge