Olivier Scheidegger
pymia: A Python package for data handling and evaluation in deep learning-based medical image analysis
Oct 07, 2020



Abstract:Background and Objective: Deep learning enables tremendous progress in medical image analysis. One driving force of this progress are open-source frameworks like TensorFlow and PyTorch. However, these frameworks rarely address issues specific to the domain of medical image analysis, such as 3-D data handling and distance metrics for evaluation. pymia, an open-source Python package, tries to address these issues by providing flexible data handling and evaluation independent of the deep learning framework. Methods: The pymia package provides data handling and evaluation functionalities. The data handling allows flexible medical image handling in every commonly used format (e.g., 2-D, 2.5-D, and 3-D; full- or patch-wise). Even data beyond images like demographics or clinical reports can easily be integrated into deep learning pipelines. The evaluation allows stand-alone result calculation and reporting, as well as performance monitoring during training using a vast amount of domain-specific metrics for segmentation, reconstruction, and regression. Results: The pymia package is highly flexible, allows for fast prototyping, and reduces the burden of implementing data handling routines and evaluation methods. While data handling and evaluation are independent of the deep learning framework used, they can easily be integrated into TensorFlow and PyTorch pipelines. The developed package was successfully used in a variety of research projects for segmentation, reconstruction, and regression. Conclusions: The pymia package fills the gap of current deep learning frameworks regarding data handling and evaluation in medical image analysis. It is available at https://github.com/rundherum/pymia and can directly be installed from the Python Package Index using pip install pymia.
Learning Bloch Simulations for MR Fingerprinting by Invertible Neural Networks
Aug 10, 2020



Abstract:Magnetic resonance fingerprinting (MRF) enables fast and multiparametric MR imaging. Despite fast acquisition, the state-of-the-art reconstruction of MRF based on dictionary matching is slow and lacks scalability. To overcome these limitations, neural network (NN) approaches estimating MR parameters from fingerprints have been proposed recently. Here, we revisit NN-based MRF reconstruction to jointly learn the forward process from MR parameters to fingerprints and the backward process from fingerprints to MR parameters by leveraging invertible neural networks (INNs). As a proof-of-concept, we perform various experiments showing the benefit of learning the forward process, i.e., the Bloch simulations, for improved MR parameter estimation. The benefit especially accentuates when MR parameter estimation is difficult due to MR physical restrictions. Therefore, INNs might be a feasible alternative to the current solely backward-based NNs for MRF reconstruction.
Spatially Regularized Parametric Map Reconstruction for Fast Magnetic Resonance Fingerprinting
Nov 09, 2019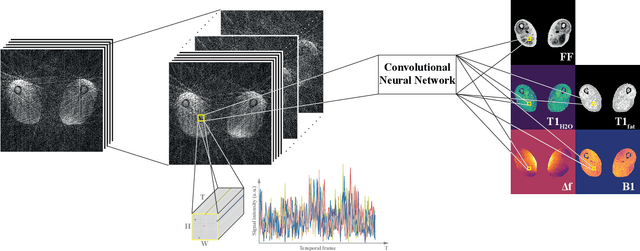


Abstract:Magnetic resonance fingerprinting (MRF) provides a unique concept for simultaneous and fast acquisition of multiple quantitative MR parameters. Despite acquisition efficiency, adoption of MRF into the clinics is hindered by its dictionary-based reconstruction, which is computationally demanding and lacks scalability. Here, we propose a convolutional neural network-based reconstruction, which enables both accurate and fast reconstruction of parametric maps, and is adaptable based on the needs of spatial regularization and the capacity for the reconstruction. We evaluated the method using MRF T1-FF, an MRF sequence for T1 relaxation time of water and fat fraction mapping. We demonstrate the method's performance on a highly heterogeneous dataset consisting of 164 patients with various neuromuscular diseases imaged at thighs and legs. We empirically show the benefit of incorporating spatial regularization during the reconstruction and demonstrate that the method learns meaningful features from MR physics perspective. Further, we investigate the ability of the method to handle highly heterogeneous morphometric variations and its generalization to anatomical regions unseen during training. The obtained results outperform the state-of-the-art in deep learning-based MRF reconstruction. Coupled with fast MRF sequences, the proposed method has the potential of enabling multiparametric MR imaging in clinically feasible time.
Learning Shape Representation on Sparse Point Clouds for Volumetric Image Segmentation
Jun 05, 2019



Abstract:Volumetric image segmentation with convolutional neural networks (CNNs) encounters several challenges, which are specific to medical images. Among these challenges are large volumes of interest, high class imbalances, and difficulties in learning shape representations. To tackle these challenges, we propose to improve over traditional CNN-based volumetric image segmentation through point-wise classification of point clouds. The sparsity of point clouds allows processing of entire image volumes, balancing highly imbalanced segmentation problems, and explicitly learning an anatomical shape. We build upon PointCNN, a neural network proposed to process point clouds, and propose here to jointly encode shape and volumetric information within the point cloud in a compact and computationally effective manner. We demonstrate how this approach can then be used to refine CNN-based segmentation, which yields significantly improved results in our experiments on the difficult task of peripheral nerve segmentation from magnetic resonance neurography images. By synthetic experiments, we further show the capability of our approach in learning an explicit anatomical shape representation.
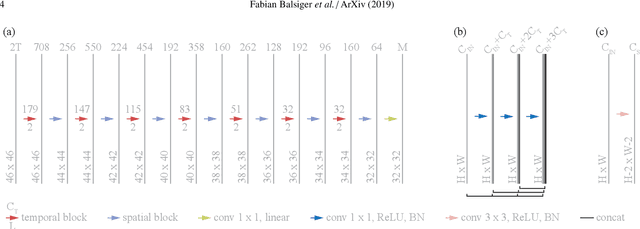
Magnetic Resonance Fingerprinting Reconstruction via Spatiotemporal Convolutional Neural Networks
Jul 24, 2018



Abstract:Magnetic resonance fingerprinting (MRF) quantifies multiple nuclear magnetic resonance parameters in a single and fast acquisition. Standard MRF reconstructs parametric maps using dictionary matching, which lacks scalability due to computational inefficiency. We propose to perform MRF map reconstruction using a spatiotemporal convolutional neural network, which exploits the relationship between neighboring MRF signal evolutions to replace the dictionary matching. We evaluate our method on multiparametric brain scans and compare it to three recent MRF reconstruction approaches. Our method achieves state-of-the-art reconstruction accuracy and yields qualitatively more appealing maps compared to other reconstruction methods. In addition, the reconstruction time is significantly reduced compared to a dictionary-based approach.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge