Yilong Li
GVGS: Gaussian Visibility-Aware Multi-View Geometry for Accurate Surface Reconstruction
Jan 28, 2026Abstract:3D Gaussian Splatting enables efficient optimization and high-quality rendering, yet accurate surface reconstruction remains challenging. Prior methods improve surface reconstruction by refining Gaussian depth estimates, either via multi-view geometric consistency or through monocular depth priors. However, multi-view constraints become unreliable under large geometric discrepancies, while monocular priors suffer from scale ambiguity and local inconsistency, ultimately leading to inaccurate Gaussian depth supervision. To address these limitations, we introduce a Gaussian visibility-aware multi-view geometric consistency constraint that aggregates the visibility of shared Gaussian primitives across views, enabling more accurate and stable geometric supervision. In addition, we propose a progressive quadtree-calibrated Monocular depth constraint that performs block-wise affine calibration from coarse to fine spatial scales, mitigating the scale ambiguity of depth priors while preserving fine-grained surface details. Extensive experiments on DTU and TNT datasets demonstrate consistent improvements in geometric accuracy over prior Gaussian-based and implicit surface reconstruction methods. Codes are available at an anonymous repository: https://github.com/GVGScode/GVGS.
Cross-Task Experiential Learning on LLM-based Multi-Agent Collaboration
May 29, 2025Abstract:Large Language Model-based multi-agent systems (MAS) have shown remarkable progress in solving complex tasks through collaborative reasoning and inter-agent critique. However, existing approaches typically treat each task in isolation, resulting in redundant computations and limited generalization across structurally similar tasks. To address this, we introduce multi-agent cross-task experiential learning (MAEL), a novel framework that endows LLM-driven agents with explicit cross-task learning and experience accumulation. We model the task-solving workflow on a graph-structured multi-agent collaboration network, where agents propagate information and coordinate via explicit connectivity. During the experiential learning phase, we quantify the quality for each step in the task-solving workflow and store the resulting rewards along with the corresponding inputs and outputs into each agent's individual experience pool. During inference, agents retrieve high-reward, task-relevant experiences as few-shot examples to enhance the effectiveness of each reasoning step, thereby enabling more accurate and efficient multi-agent collaboration. Experimental results on diverse datasets demonstrate that MAEL empowers agents to learn from prior task experiences effectively-achieving faster convergence and producing higher-quality solutions on current tasks.
FL-PLAS: Federated Learning with Partial Layer Aggregation for Backdoor Defense Against High-Ratio Malicious Clients
May 17, 2025Abstract:Federated learning (FL) is gaining increasing attention as an emerging collaborative machine learning approach, particularly in the context of large-scale computing and data systems. However, the fundamental algorithm of FL, Federated Averaging (FedAvg), is susceptible to backdoor attacks. Although researchers have proposed numerous defense algorithms, two significant challenges remain. The attack is becoming more stealthy and harder to detect, and current defense methods are unable to handle 50\% or more malicious users or assume an auxiliary server dataset. To address these challenges, we propose a novel defense algorithm, FL-PLAS, \textbf{F}ederated \textbf{L}earning based on \textbf{P}artial\textbf{ L}ayer \textbf{A}ggregation \textbf{S}trategy. In particular, we divide the local model into a feature extractor and a classifier. In each iteration, the clients only upload the parameters of a feature extractor after local training. The server then aggregates these local parameters and returns the results to the clients. Each client retains its own classifier layer, ensuring that the backdoor labels do not impact other clients. We assess the effectiveness of FL-PLAS against state-of-the-art (SOTA) backdoor attacks on three image datasets and compare our approach to six defense strategies. The results of the experiment demonstrate that our methods can effectively protect local models from backdoor attacks. Without requiring any auxiliary dataset for the server, our method achieves a high main-task accuracy with a lower backdoor accuracy even under the condition of 90\% malicious users with the attacks of trigger, semantic and edge-case.
HUG: Hierarchical Urban Gaussian Splatting with Block-Based Reconstruction
Apr 23, 2025Abstract:As urban 3D scenes become increasingly complex and the demand for high-quality rendering grows, efficient scene reconstruction and rendering techniques become crucial. We present HUG, a novel approach to address inefficiencies in handling large-scale urban environments and intricate details based on 3D Gaussian splatting. Our method optimizes data partitioning and the reconstruction pipeline by incorporating a hierarchical neural Gaussian representation. We employ an enhanced block-based reconstruction pipeline focusing on improving reconstruction quality within each block and reducing the need for redundant training regions around block boundaries. By integrating neural Gaussian representation with a hierarchical architecture, we achieve high-quality scene rendering at a low computational cost. This is demonstrated by our state-of-the-art results on public benchmarks, which prove the effectiveness and advantages in large-scale urban scene representation.
PalmBench: A Comprehensive Benchmark of Compressed Large Language Models on Mobile Platforms
Oct 05, 2024



Abstract:Deploying large language models (LLMs) locally on mobile devices is advantageous in scenarios where transmitting data to remote cloud servers is either undesirable due to privacy concerns or impractical due to network connection. Recent advancements (MLC, 2023a; Gerganov, 2023) have facilitated the local deployment of LLMs. However, local deployment also presents challenges, particularly in balancing quality (generative performance), latency, and throughput within the hardware constraints of mobile devices. In this paper, we introduce our lightweight, all-in-one automated benchmarking framework that allows users to evaluate LLMs on mobile devices. We provide a comprehensive benchmark of various popular LLMs with different quantization configurations (both weights and activations) across multiple mobile platforms with varying hardware capabilities. Unlike traditional benchmarks that assess full-scale models on high-end GPU clusters, we focus on evaluating resource efficiency (memory and power consumption) and harmful output for compressed models on mobile devices. Our key observations include i) differences in energy efficiency and throughput across mobile platforms; ii) the impact of quantization on memory usage, GPU execution time, and power consumption; and iii) accuracy and performance degradation of quantized models compared to their non-quantized counterparts; and iv) the frequency of hallucinations and toxic content generated by compressed LLMs on mobile devices.
Neural Laplacian Operator for 3D Point Clouds
Sep 10, 2024



Abstract:The discrete Laplacian operator holds a crucial role in 3D geometry processing, yet it is still challenging to define it on point clouds. Previous works mainly focused on constructing a local triangulation around each point to approximate the underlying manifold for defining the Laplacian operator, which may not be robust or accurate. In contrast, we simply use the K-nearest neighbors (KNN) graph constructed from the input point cloud and learn the Laplacian operator on the KNN graph with graph neural networks (GNNs). However, the ground-truth Laplacian operator is defined on a manifold mesh with a different connectivity from the KNN graph and thus cannot be directly used for training. To train the GNN, we propose a novel training scheme by imitating the behavior of the ground-truth Laplacian operator on a set of probe functions so that the learned Laplacian operator behaves similarly to the ground-truth Laplacian operator. We train our network on a subset of ShapeNet and evaluate it across a variety of point clouds. Compared with previous methods, our method reduces the error by an order of magnitude and excels in handling sparse point clouds with thin structures or sharp features. Our method also demonstrates a strong generalization ability to unseen shapes. With our learned Laplacian operator, we further apply a series of Laplacian-based geometry processing algorithms directly to point clouds and achieve accurate results, enabling many exciting possibilities for geometry processing on point clouds. The code and trained models are available at https://github.com/IntelligentGeometry/NeLo.
Gemini: Integrating Full-fledged Sensing upon Millimeter Wave Communications
Jul 04, 2024
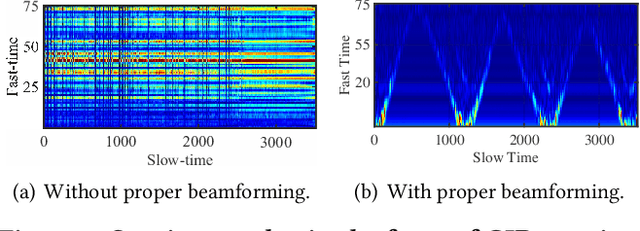
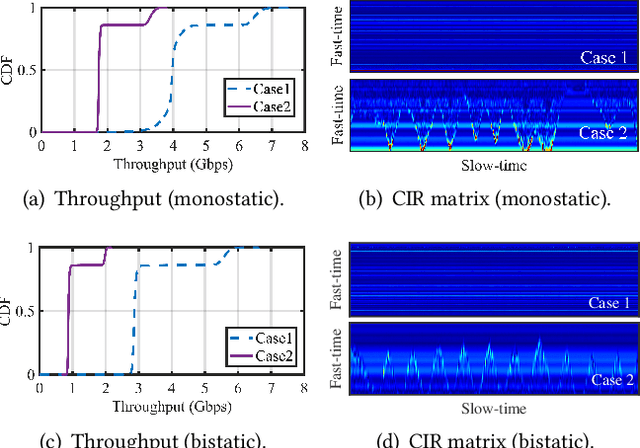
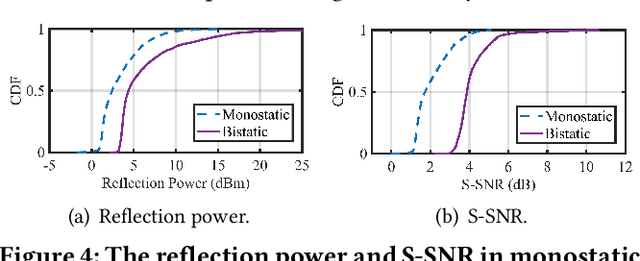
Abstract:Integrating millimeter wave (mmWave)technology in both communication and sensing is promising as it enables the reuse of existing spectrum and infrastructure without draining resources. Most existing systems piggyback sensing onto conventional communication modes without fully exploiting the potential of integrated sensing and communication (ISAC) in mmWave radios (not full-fledged). In this paper, we design and implement a full-fledged mmWave ISAC system Gemini; it delivers raw channel states to serve a broad category of sensing applications. We first propose the mmWave self-interference cancellation approach to extract the weak reflected signals for near-field sensing purposes. Then, we develop a joint optimization scheduling framework that can be utilized in accurate radar sensing while maximizing the communication throughput. Finally, we design a united fusion sensing algorithm to offer a better sensing performance via combining monostatic and bistatic modes. We evaluate our system in extensive experiments to demonstrate Gemini's capability of simultaneously operating sensing and communication, enabling mmWave ISAC to perform better than the commercial off-the-shelf mmWave radar for 5G cellular networks.
MEDUSA: Scalable Biometric Sensing in the Wild through Distributed MIMO Radars
Oct 10, 2023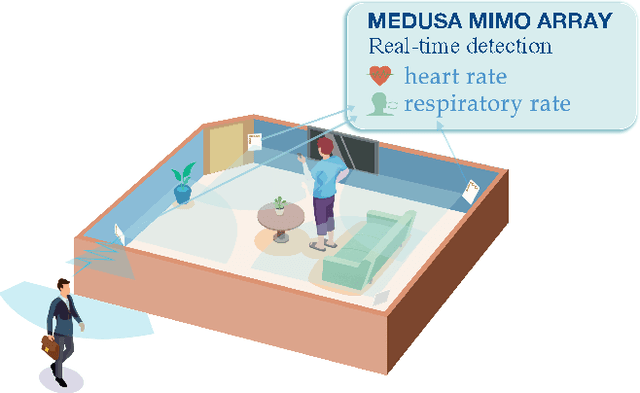
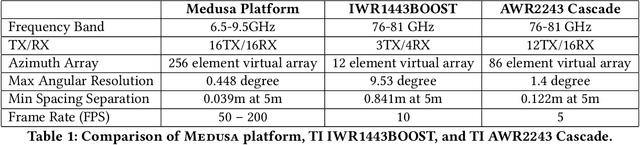
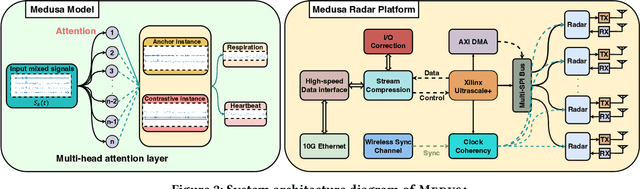
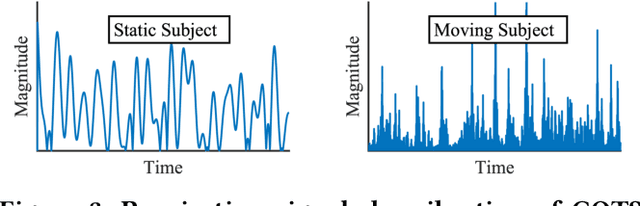
Abstract:Radar-based techniques for detecting vital signs have shown promise for continuous contactless vital sign sensing and healthcare applications. However, real-world indoor environments face significant challenges for existing vital sign monitoring systems. These include signal blockage in non-line-of-sight (NLOS) situations, movement of human subjects, and alterations in location and orientation. Additionally, these existing systems failed to address the challenge of tracking multiple targets simultaneously. To overcome these challenges, we present MEDUSA, a novel coherent ultra-wideband (UWB) based distributed multiple-input multiple-output (MIMO) radar system, especially it allows users to customize and disperse the $16 \times 16$ into sub-arrays. MEDUSA takes advantage of the diversity benefits of distributed yet wirelessly synchronized MIMO arrays to enable robust vital sign monitoring in real-world and daily living environments where human targets are moving and surrounded by obstacles. We've developed a scalable, self-supervised contrastive learning model which integrates seamlessly with our hardware platform. Each attention weight within the model corresponds to a specific antenna pair of Tx and Rx. The model proficiently recovers accurate vital sign waveforms by decomposing and correlating the mixed received signals, including comprising human motion, mobility, noise, and vital signs. Through extensive evaluations involving 21 participants and over 200 hours of collected data (3.75 TB in total, with 1.89 TB for static subjects and 1.86 TB for moving subjects), MEDUSA's performance has been validated, showing an average gain of 20% compared to existing systems employing COTS radar sensors. This demonstrates MEDUSA's spatial diversity gain for real-world vital sign monitoring, encompassing target and environmental dynamics in familiar and unfamiliar indoor environments.
CTooth+: A Large-scale Dental Cone Beam Computed Tomography Dataset and Benchmark for Tooth Volume Segmentation
Aug 02, 2022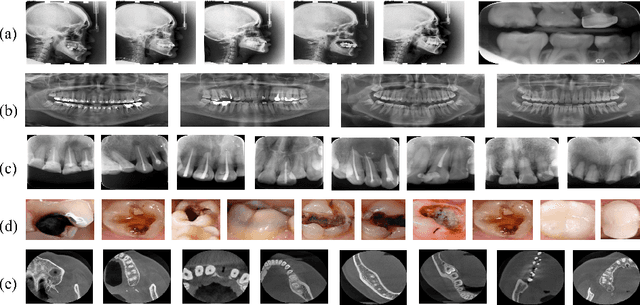
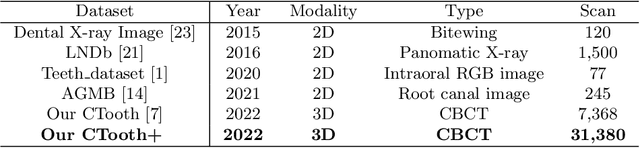
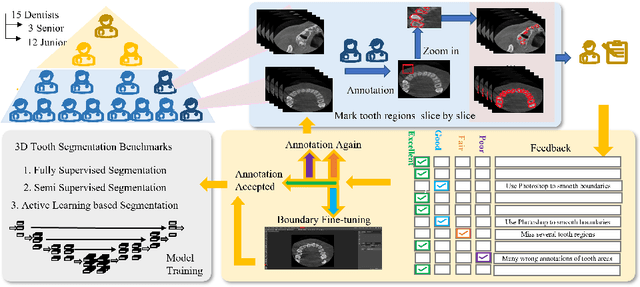
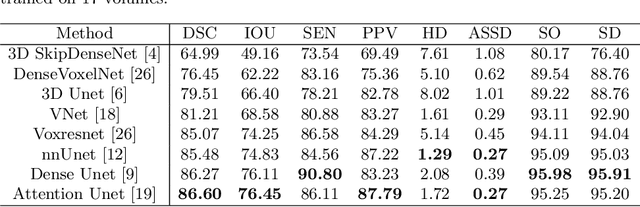
Abstract:Accurate tooth volume segmentation is a prerequisite for computer-aided dental analysis. Deep learning-based tooth segmentation methods have achieved satisfying performances but require a large quantity of tooth data with ground truth. The dental data publicly available is limited meaning the existing methods can not be reproduced, evaluated and applied in clinical practice. In this paper, we establish a 3D dental CBCT dataset CTooth+, with 22 fully annotated volumes and 146 unlabeled volumes. We further evaluate several state-of-the-art tooth volume segmentation strategies based on fully-supervised learning, semi-supervised learning and active learning, and define the performance principles. This work provides a new benchmark for the tooth volume segmentation task, and the experiment can serve as the baseline for future AI-based dental imaging research and clinical application development.
Evaluating the Practicality of Learned Image Compression
Jul 29, 2022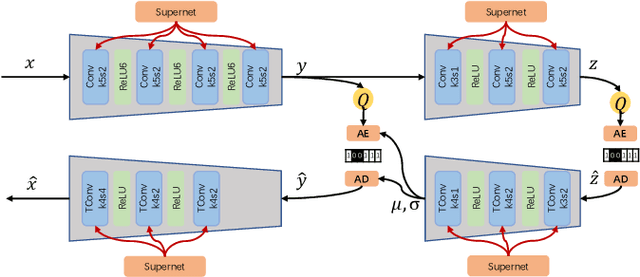
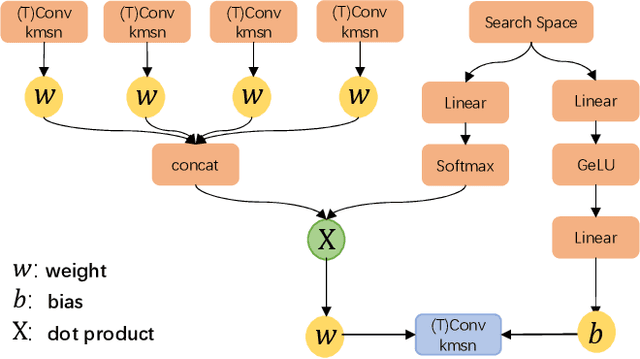
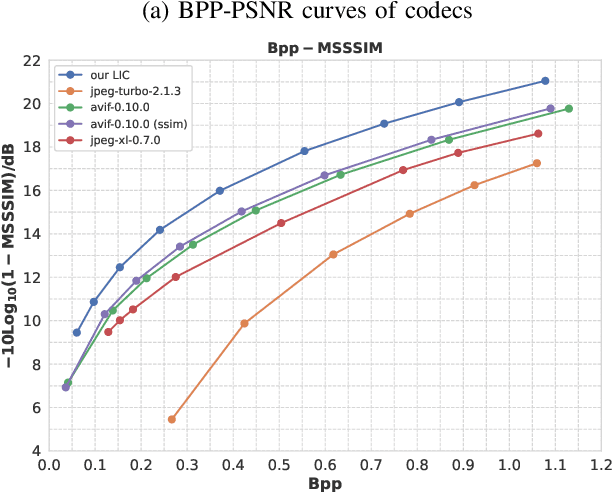
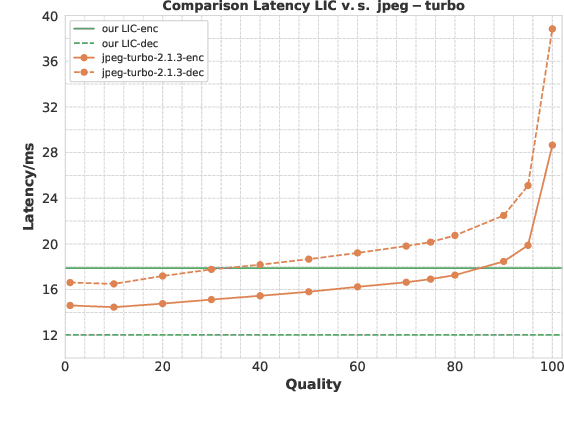
Abstract:Learned image compression has achieved extraordinary rate-distortion performance in PSNR and MS-SSIM compared to traditional methods. However, it suffers from intensive computation, which is intolerable for real-world applications and leads to its limited industrial application for now. In this paper, we introduce neural architecture search (NAS) to designing more efficient networks with lower latency, and leverage quantization to accelerate the inference process. Meanwhile, efforts in engineering like multi-threading and SIMD have been made to improve efficiency. Optimized using a hybrid loss of PSNR and MS-SSIM for better visual quality, we obtain much higher MS-SSIM than JPEG, JPEG XL and AVIF over all bit rates, and PSNR between that of JPEG XL and AVIF. Our software implementation of LIC achieves comparable or even faster inference speed compared to jpeg-turbo while being multiple times faster than JPEG XL and AVIF. Besides, our implementation of LIC reaches stunning throughput of 145 fps for encoding and 208 fps for decoding on a Tesla T4 GPU for 1080p images. On CPU, the latency of our implementation is comparable with JPEG XL.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge