Lihong Liu
HumanDreamer-X: Photorealistic Single-image Human Avatars Reconstruction via Gaussian Restoration
Apr 04, 2025
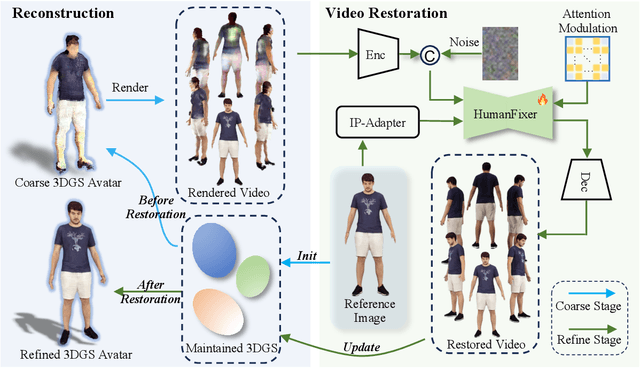

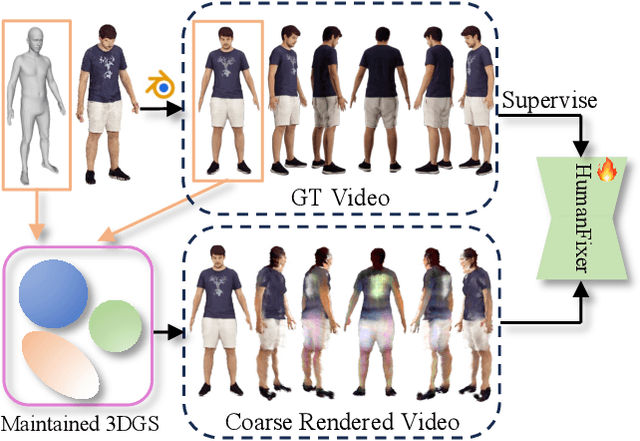
Abstract:Single-image human reconstruction is vital for digital human modeling applications but remains an extremely challenging task. Current approaches rely on generative models to synthesize multi-view images for subsequent 3D reconstruction and animation. However, directly generating multiple views from a single human image suffers from geometric inconsistencies, resulting in issues like fragmented or blurred limbs in the reconstructed models. To tackle these limitations, we introduce \textbf{HumanDreamer-X}, a novel framework that integrates multi-view human generation and reconstruction into a unified pipeline, which significantly enhances the geometric consistency and visual fidelity of the reconstructed 3D models. In this framework, 3D Gaussian Splatting serves as an explicit 3D representation to provide initial geometry and appearance priority. Building upon this foundation, \textbf{HumanFixer} is trained to restore 3DGS renderings, which guarantee photorealistic results. Furthermore, we delve into the inherent challenges associated with attention mechanisms in multi-view human generation, and propose an attention modulation strategy that effectively enhances geometric details identity consistency across multi-view. Experimental results demonstrate that our approach markedly improves generation and reconstruction PSNR quality metrics by 16.45% and 12.65%, respectively, achieving a PSNR of up to 25.62 dB, while also showing generalization capabilities on in-the-wild data and applicability to various human reconstruction backbone models.
HumanDreamer: Generating Controllable Human-Motion Videos via Decoupled Generation
Apr 01, 2025Abstract:Human-motion video generation has been a challenging task, primarily due to the difficulty inherent in learning human body movements. While some approaches have attempted to drive human-centric video generation explicitly through pose control, these methods typically rely on poses derived from existing videos, thereby lacking flexibility. To address this, we propose HumanDreamer, a decoupled human video generation framework that first generates diverse poses from text prompts and then leverages these poses to generate human-motion videos. Specifically, we propose MotionVid, the largest dataset for human-motion pose generation. Based on the dataset, we present MotionDiT, which is trained to generate structured human-motion poses from text prompts. Besides, a novel LAMA loss is introduced, which together contribute to a significant improvement in FID by 62.4%, along with respective enhancements in R-precision for top1, top2, and top3 by 41.8%, 26.3%, and 18.3%, thereby advancing both the Text-to-Pose control accuracy and FID metrics. Our experiments across various Pose-to-Video baselines demonstrate that the poses generated by our method can produce diverse and high-quality human-motion videos. Furthermore, our model can facilitate other downstream tasks, such as pose sequence prediction and 2D-3D motion lifting.
Dynamic Evidence Decoupling for Trusted Multi-view Learning
Oct 04, 2024



Abstract:Multi-view learning methods often focus on improving decision accuracy, while neglecting the decision uncertainty, limiting their suitability for safety-critical applications. To mitigate this, researchers propose trusted multi-view learning methods that estimate classification probabilities and uncertainty by learning the class distributions for each instance. However, these methods assume that the data from each view can effectively differentiate all categories, ignoring the semantic vagueness phenomenon in real-world multi-view data. Our findings demonstrate that this phenomenon significantly suppresses the learning of view-specific evidence in existing methods. We propose a Consistent and Complementary-aware trusted Multi-view Learning (CCML) method to solve this problem. We first construct view opinions using evidential deep neural networks, which consist of belief mass vectors and uncertainty estimates. Next, we dynamically decouple the consistent and complementary evidence. The consistent evidence is derived from the shared portions across all views, while the complementary evidence is obtained by averaging the differing portions across all views. We ensure that the opinion constructed from the consistent evidence strictly aligns with the ground-truth category. For the opinion constructed from the complementary evidence, we allow it for potential vagueness in the evidence. We compare CCML with state-of-the-art baselines on one synthetic and six real-world datasets. The results validate the effectiveness of the dynamic evidence decoupling strategy and show that CCML significantly outperforms baselines on accuracy and reliability. The code is released at https://github.com/Lihong-Liu/CCML.
Deep Learning Methods for Lung Cancer Segmentation in Whole-slide Histopathology Images -- the ACDC@LungHP Challenge 2019
Aug 21, 2020
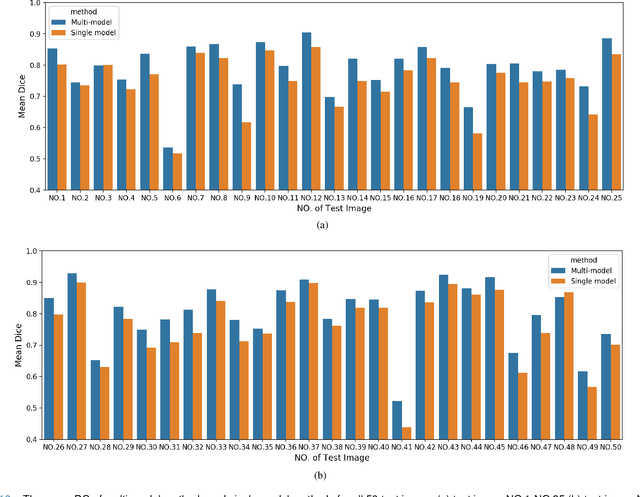
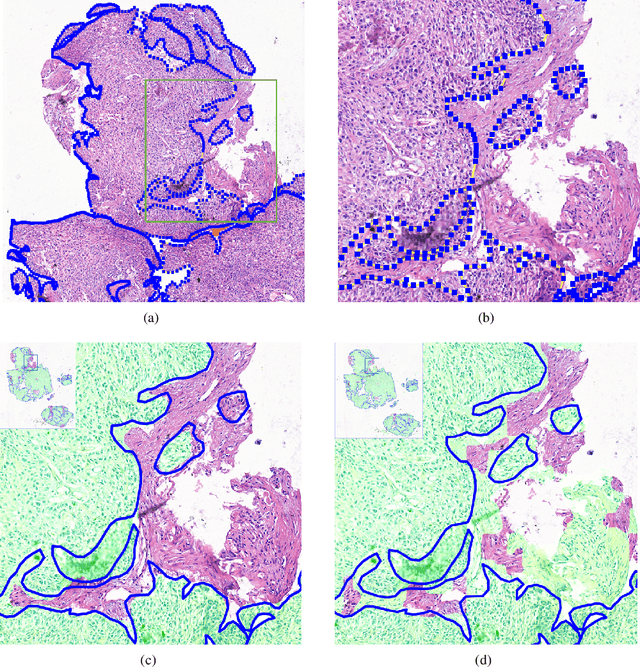
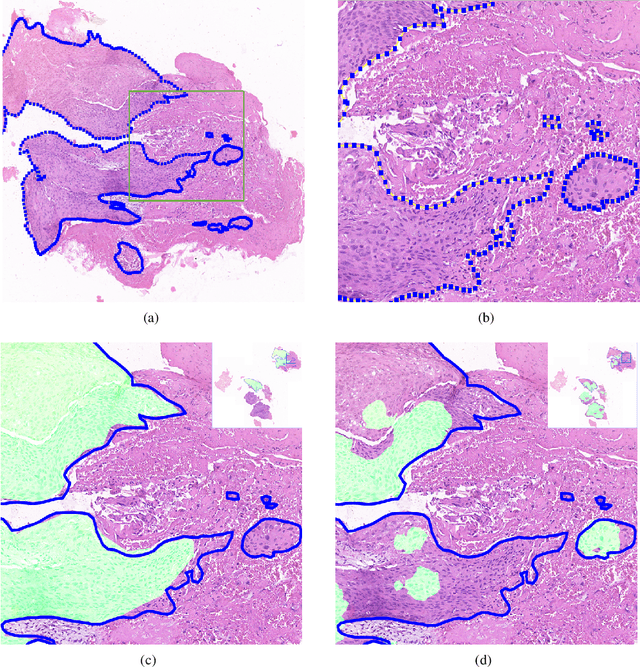
Abstract:Accurate segmentation of lung cancer in pathology slides is a critical step in improving patient care. We proposed the ACDC@LungHP (Automatic Cancer Detection and Classification in Whole-slide Lung Histopathology) challenge for evaluating different computer-aided diagnosis (CADs) methods on the automatic diagnosis of lung cancer. The ACDC@LungHP 2019 focused on segmentation (pixel-wise detection) of cancer tissue in whole slide imaging (WSI), using an annotated dataset of 150 training images and 50 test images from 200 patients. This paper reviews this challenge and summarizes the top 10 submitted methods for lung cancer segmentation. All methods were evaluated using the false positive rate, false negative rate, and DICE coefficient (DC). The DC ranged from 0.7354$\pm$0.1149 to 0.8372$\pm$0.0858. The DC of the best method was close to the inter-observer agreement (0.8398$\pm$0.0890). All methods were based on deep learning and categorized into two groups: multi-model method and single model method. In general, multi-model methods were significantly better ($\textit{p}$<$0.01$) than single model methods, with mean DC of 0.7966 and 0.7544, respectively. Deep learning based methods could potentially help pathologists find suspicious regions for further analysis of lung cancer in WSI.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge