Xichao Teng
3MOS: Multi-sources, Multi-resolutions, and Multi-scenes dataset for Optical-SAR image matching
Apr 01, 2024



Abstract:Optical-SAR image matching is a fundamental task for image fusion and visual navigation. However, all large-scale open SAR dataset for methods development are collected from single platform, resulting in limited satellite types and spatial resolutions. Since images captured by different sensors vary significantly in both geometric and radiometric appearance, existing methods may fail to match corresponding regions containing the same content. Besides, most of existing datasets have not been categorized based on the characteristics of different scenes. To encourage the design of more general multi-modal image matching methods, we introduce a large-scale Multi-sources,Multi-resolutions, and Multi-scenes dataset for Optical-SAR image matching(3MOS). It consists of 155K optical-SAR image pairs, including SAR data from six commercial satellites, with resolutions ranging from 1.25m to 12.5m. The data has been classified into eight scenes including urban, rural, plains, hills, mountains, water, desert, and frozen earth. Extensively experiments show that none of state-of-the-art methods achieve consistently superior performance across different sources, resolutions and scenes. In addition, the distribution of data has a substantial impact on the matching capability of deep learning models, this proposes the domain adaptation challenge in optical-SAR image matching. Our data and code will be available at:https://github.com/3M-OS/3MOS.
Deep Learning Methods for Lung Cancer Segmentation in Whole-slide Histopathology Images -- the ACDC@LungHP Challenge 2019
Aug 21, 2020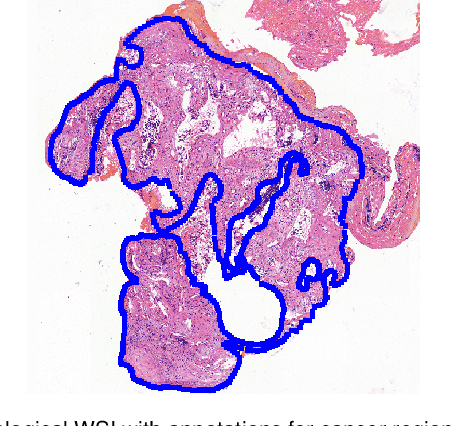
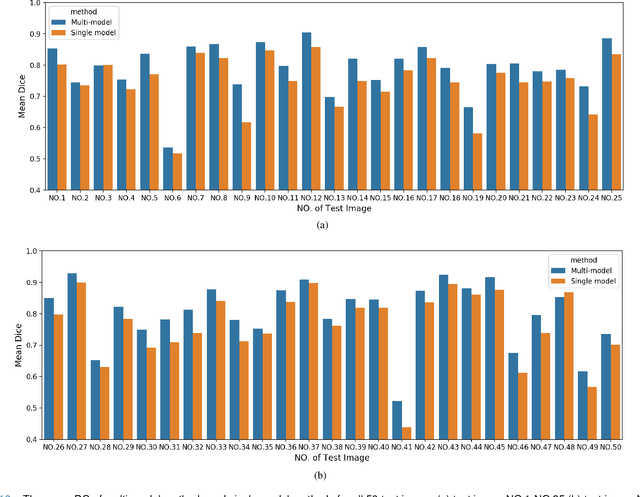
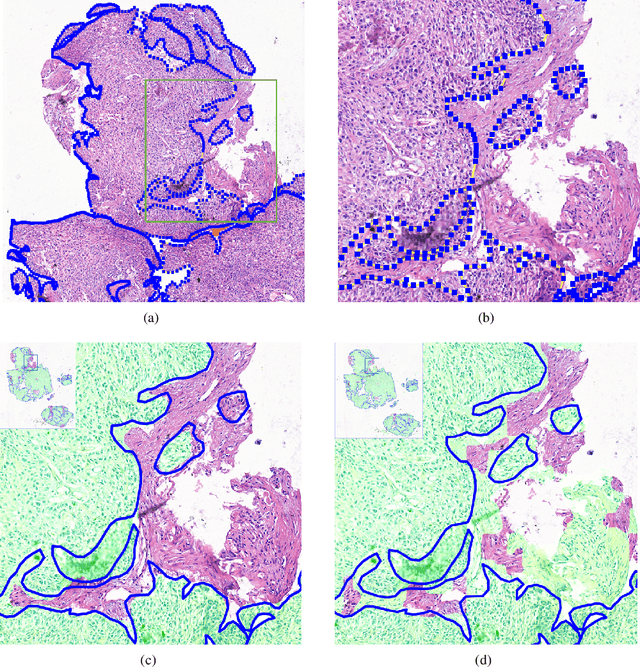
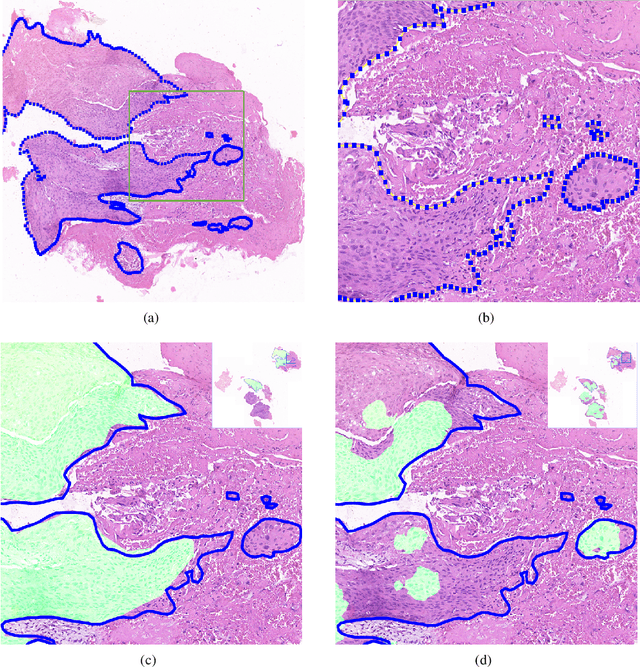
Abstract:Accurate segmentation of lung cancer in pathology slides is a critical step in improving patient care. We proposed the ACDC@LungHP (Automatic Cancer Detection and Classification in Whole-slide Lung Histopathology) challenge for evaluating different computer-aided diagnosis (CADs) methods on the automatic diagnosis of lung cancer. The ACDC@LungHP 2019 focused on segmentation (pixel-wise detection) of cancer tissue in whole slide imaging (WSI), using an annotated dataset of 150 training images and 50 test images from 200 patients. This paper reviews this challenge and summarizes the top 10 submitted methods for lung cancer segmentation. All methods were evaluated using the false positive rate, false negative rate, and DICE coefficient (DC). The DC ranged from 0.7354$\pm$0.1149 to 0.8372$\pm$0.0858. The DC of the best method was close to the inter-observer agreement (0.8398$\pm$0.0890). All methods were based on deep learning and categorized into two groups: multi-model method and single model method. In general, multi-model methods were significantly better ($\textit{p}$<$0.01$) than single model methods, with mean DC of 0.7966 and 0.7544, respectively. Deep learning based methods could potentially help pathologists find suspicious regions for further analysis of lung cancer in WSI.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge