Steffen E. Petersen
National Institute for Health Research, Barts Heart Centre, St Bartholomew's Hospital, Barts Health NHS Trust, London, UK, Health Data Research UK, London, UK, Alan Turing Institute, London, UK
SAGCNet: Spatial-Aware Graph Completion Network for Missing Slice Imputation in Population CMR Imaging
Aug 09, 2025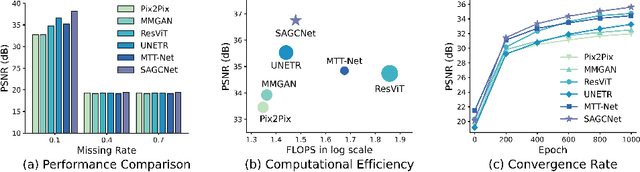
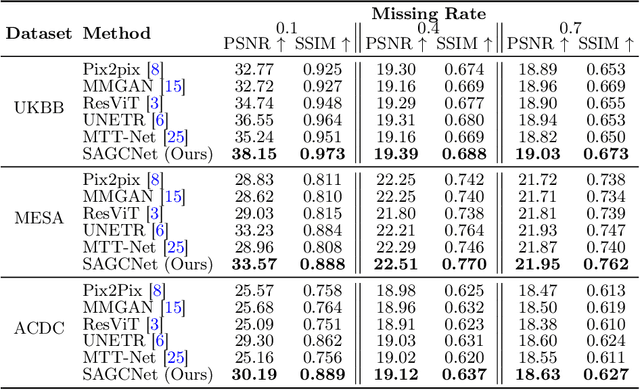
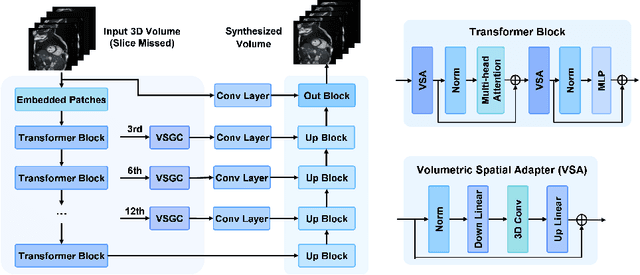
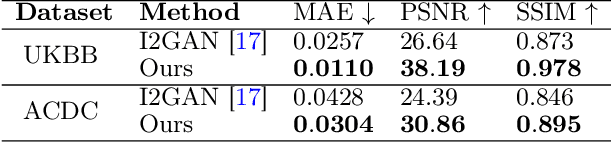
Abstract:Magnetic resonance imaging (MRI) provides detailed soft-tissue characteristics that assist in disease diagnosis and screening. However, the accuracy of clinical practice is often hindered by missing or unusable slices due to various factors. Volumetric MRI synthesis methods have been developed to address this issue by imputing missing slices from available ones. The inherent 3D nature of volumetric MRI data, such as cardiac magnetic resonance (CMR), poses significant challenges for missing slice imputation approaches, including (1) the difficulty of modeling local inter-slice correlations and dependencies of volumetric slices, and (2) the limited exploration of crucial 3D spatial information and global context. In this study, to mitigate these issues, we present Spatial-Aware Graph Completion Network (SAGCNet) to overcome the dependency on complete volumetric data, featuring two main innovations: (1) a volumetric slice graph completion module that incorporates the inter-slice relationships into a graph structure, and (2) a volumetric spatial adapter component that enables our model to effectively capture and utilize various forms of 3D spatial context. Extensive experiments on cardiac MRI datasets demonstrate that SAGCNet is capable of synthesizing absent CMR slices, outperforming competitive state-of-the-art MRI synthesis methods both quantitatively and qualitatively. Notably, our model maintains superior performance even with limited slice data.
Commentary on explainable artificial intelligence methods: SHAP and LIME
May 08, 2023Abstract:eXplainable artificial intelligence (XAI) methods have emerged to convert the black box of machine learning models into a more digestible form. These methods help to communicate how the model works with the aim of making machine learning models more transparent and increasing the trust of end-users into their output. SHapley Additive exPlanations (SHAP) and Local Interpretable Model Agnostic Explanation (LIME) are two widely used XAI methods particularly with tabular data. In this commentary piece, we discuss the way the explainability metrics of these two methods are generated and propose a framework for interpretation of their outputs, highlighting their weaknesses and strengths.
Characterizing the contribution of dependent features in XAI methods
Apr 04, 2023Abstract:Explainable Artificial Intelligence (XAI) provides tools to help understanding how the machine learning models work and reach a specific outcome. It helps to increase the interpretability of models and makes the models more trustworthy and transparent. In this context, many XAI methods were proposed being SHAP and LIME the most popular. However, the proposed methods assume that used predictors in the machine learning models are independent which in general is not necessarily true. Such assumption casts shadows on the robustness of the XAI outcomes such as the list of informative predictors. Here, we propose a simple, yet useful proxy that modifies the outcome of any XAI feature ranking method allowing to account for the dependency among the predictors. The proposed approach has the advantage of being model-agnostic as well as simple to calculate the impact of each predictor in the model in presence of collinearity.
Large-scale, multi-centre, multi-disease validation of an AI clinical tool for cine CMR analysis
Jun 15, 2022



Abstract:INTRODUCTION: Artificial intelligence (AI) has the potential to facilitate the automation of CMR analysis for biomarker extraction. However, most AI algorithms are trained on a specific input domain (e.g., single scanner vendor or hospital-tailored imaging protocol) and lack the robustness to perform optimally when applied to CMR data from other input domains. METHODS: Our proposed framework consists of an AI-based algorithm for biventricular segmentation of short-axis images, followed by a post-analysis quality control to detect erroneous results. The segmentation algorithm was trained on a large dataset of clinical CMR scans from two NHS hospitals (n=2793) and validated on additional cases from this dataset (n=441) and on five external datasets (n=6808). The validation data included CMR scans of patients with a range of diseases acquired at 12 different centres using CMR scanners from all major vendors. RESULTS: Our method yielded median Dice scores over 87%, translating into median absolute errors in cardiac biomarkers within the range of inter-observer variability: <8.4mL (left ventricle), <9.2mL (right ventricle), <13.3g (left ventricular mass), and <5.9% (ejection fraction) across all datasets. Stratification of cases according to phenotypes of cardiac disease and scanner vendors showed good agreement. CONCLUSIONS: We show that our proposed tool, which combines a state-of-the-art AI algorithm trained on a large-scale multi-domain CMR dataset with a post-analysis quality control, allows us to robustly deal with routine clinical data from multiple centres, vendors, and cardiac diseases. This is a fundamental step for the clinical translation of AI algorithms. Moreover, our method yields a range of additional biomarkers of cardiac function (filling and ejection rates, regional wall motion, and strain) at no extra computational cost.
Fairness in Cardiac MR Image Analysis: An Investigation of Bias Due to Data Imbalance in Deep Learning Based Segmentation
Jul 01, 2021
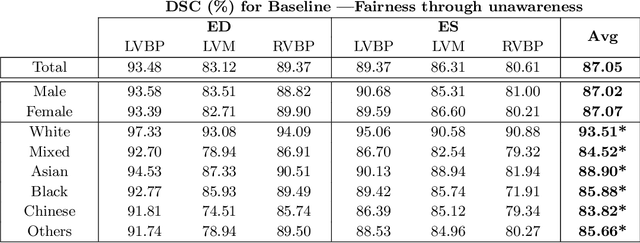

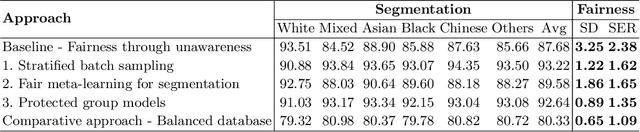
Abstract:The subject of "fairness" in artificial intelligence (AI) refers to assessing AI algorithms for potential bias based on demographic characteristics such as race and gender, and the development of algorithms to address this bias. Most applications to date have been in computer vision, although some work in healthcare has started to emerge. The use of deep learning (DL) in cardiac MR segmentation has led to impressive results in recent years, and such techniques are starting to be translated into clinical practice. However, no work has yet investigated the fairness of such models. In this work, we perform such an analysis for racial/gender groups, focusing on the problem of training data imbalance, using a nnU-Net model trained and evaluated on cine short axis cardiac MR data from the UK Biobank dataset, consisting of 5,903 subjects from 6 different racial groups. We find statistically significant differences in Dice performance between different racial groups. To reduce the racial bias, we investigated three strategies: (1) stratified batch sampling, in which batch sampling is stratified to ensure balance between racial groups; (2) fair meta-learning for segmentation, in which a DL classifier is trained to classify race and jointly optimized with the segmentation model; and (3) protected group models, in which a different segmentation model is trained for each racial group. We also compared the results to the scenario where we have a perfectly balanced database. To assess fairness we used the standard deviation (SD) and skewed error ratio (SER) of the average Dice values. Our results demonstrate that the racial bias results from the use of imbalanced training data, and that all proposed bias mitigation strategies improved fairness, with the best SD and SER resulting from the use of protected group models.
Estimating Uncertainty in Neural Networks for Cardiac MRI Segmentation: A Benchmark Study
Dec 31, 2020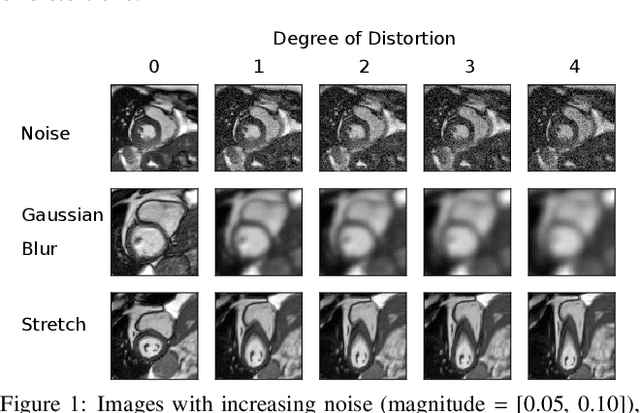
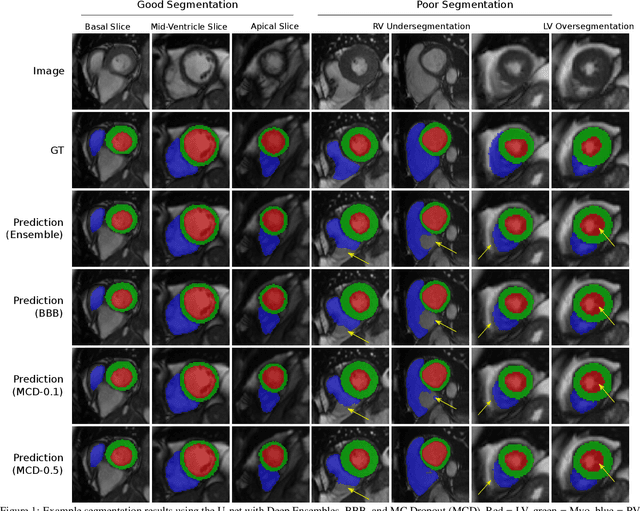
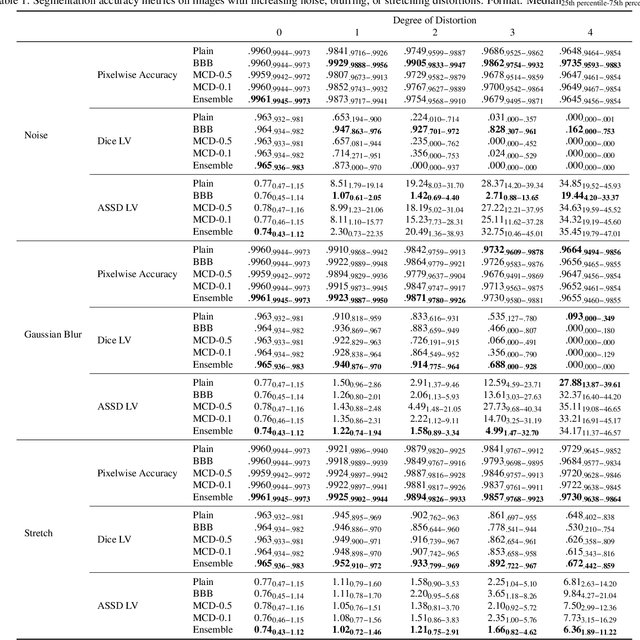
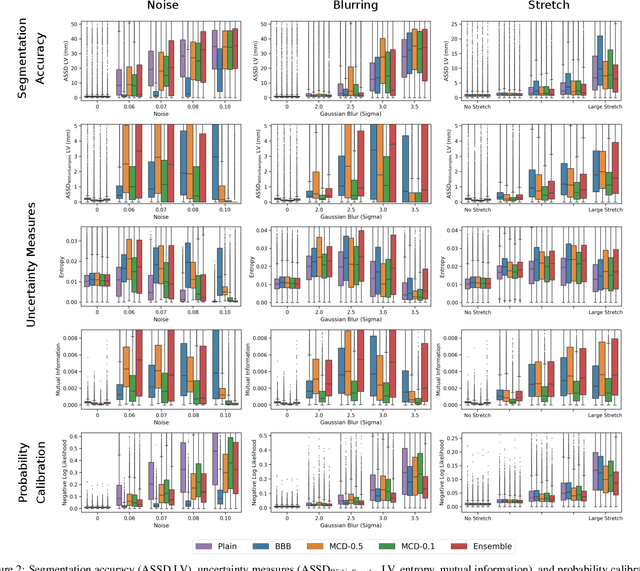
Abstract:Convolutional neural networks (CNNs) have demonstrated promise in automated cardiac magnetic resonance imaging segmentation. However, when using CNNs in a large real world dataset, it is important to quantify segmentation uncertainty in order to know which segmentations could be problematic. In this work, we performed a systematic study of Bayesian and non-Bayesian methods for estimating uncertainty in segmentation neural networks. We evaluated Bayes by Backprop (BBB), Monte Carlo (MC) Dropout, and Deep Ensembles in terms of segmentation accuracy, probability calibration, uncertainty on out-of-distribution images, and segmentation quality control. We tested these algorithms on datasets with various distortions and observed that Deep Ensembles outperformed the other methods except for images with heavy noise distortions. For segmentation quality control, we showed that segmentation uncertainty is correlated with segmentation accuracy. With the incorporation of uncertainty estimates, we were able to reduce the percentage of poor segmentation to 5% by flagging 31% to 48% of the most uncertain images for manual review, substantially lower than random review of the results without using neural network uncertainty.
A radiomics approach to analyze cardiac alterations in hypertension
Jul 21, 2020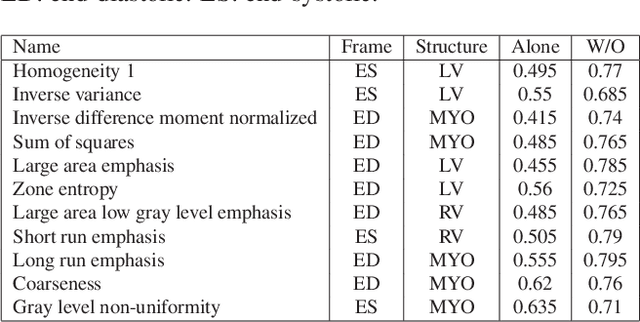
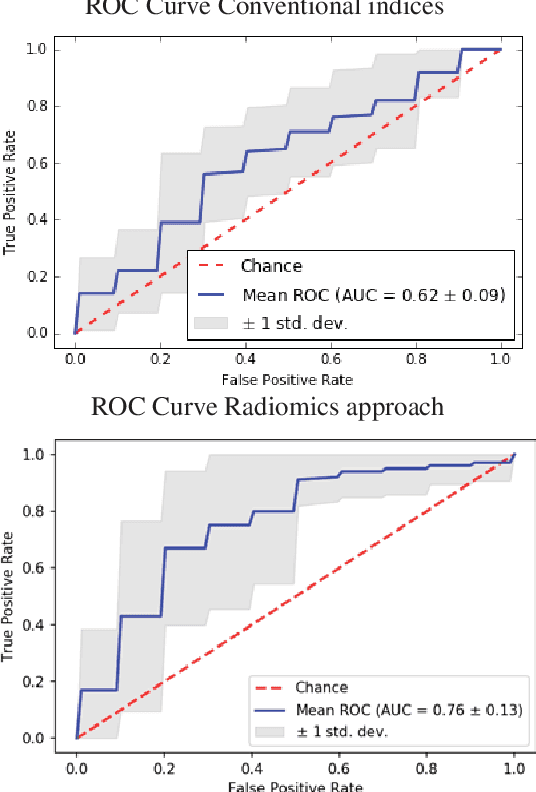
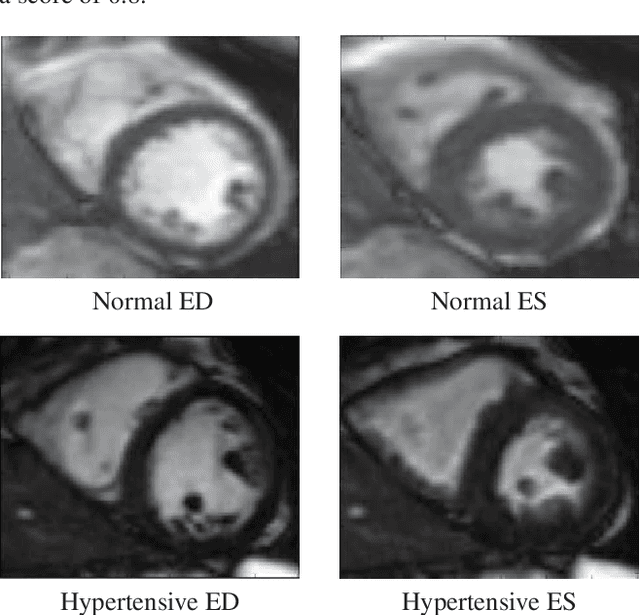
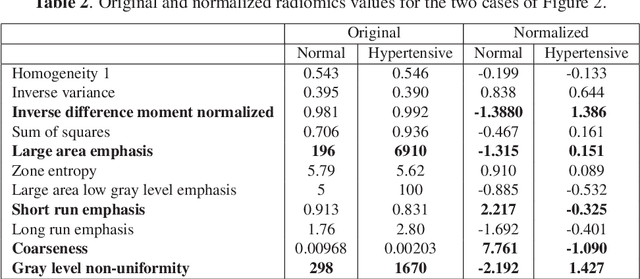
Abstract:Hypertension is a medical condition that is well-established as a risk factor for many major diseases. For example, it can cause alterations in the cardiac structure and function over time that can lead to heart related morbidity and mortality. However, at the subclinical stage, these changes are subtle and cannot be easily captured using conventional cardiovascular indices calculated from clinical cardiac imaging. In this paper, we describe a radiomics approach for identifying intermediate imaging phenotypes associated with hypertension. The method combines feature selection and machine learning techniques to identify the most subtle as well as complex structural and tissue changes in hypertensive subgroups as compared to healthy individuals. Validation based on a sample of asymptomatic hearts that include both hypertensive and non-hypertensive cases demonstrate that the proposed radiomics model is capable of detecting intensity and textural changes well beyond the capabilities of conventional imaging phenotypes, indicating its potential for improved understanding of the longitudinal effects of hypertension on cardiovascular health and disease.
A Radiomics Approach to Computer-Aided Diagnosis with Cardiac Cine-MRI
Sep 25, 2019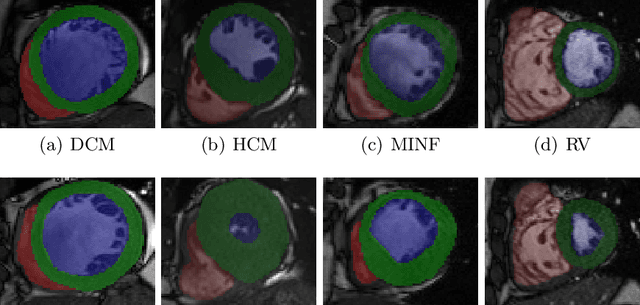
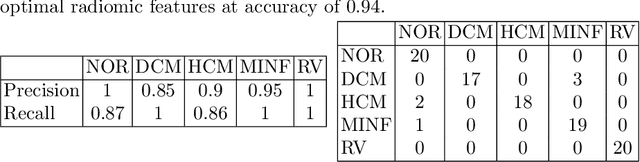
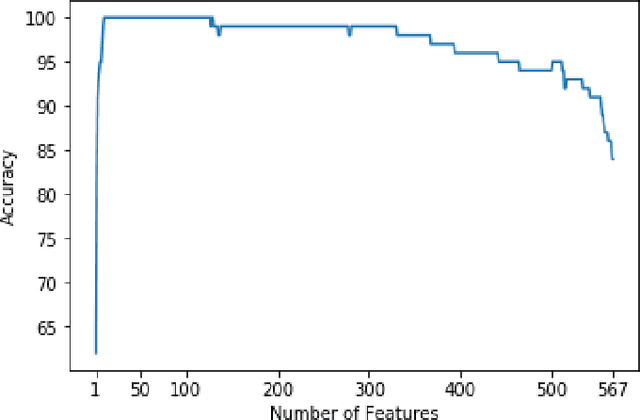
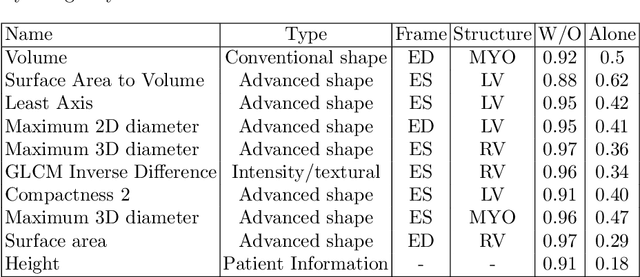
Abstract:Use expert visualization or conventional clinical indices can lack accuracy for borderline classications. Advanced statistical approaches based on eigen-decomposition have been mostly concerned with shape and motion indices. In this paper, we present a new approach to identify CVDs from cine-MRI by estimating large pools of radiomic features (statistical, shape and textural features) encoding relevant changes in anatomical and image characteristics due to CVDs. The calculated cine-MRI radiomic features are assessed using sequential forward feature selection to identify the most relevant ones for given CVD classes (e.g. myocardial infarction, cardiomyopathy, abnormal right ventricle). Finally, advanced machine learning is applied to suitably integrate the selected radiomics for final multi-feature classification based on Support Vector Machines (SVMs). The proposed technique was trained and cross-validated using 100 cine-MRI cases corresponding to five different cardiac classes from the ACDC MICCAI 2017 challenge \footnote{https://www.creatis.insa-lyon.fr/Challenge/acdc/index.html}. All cases were correctly classified in this preliminary study, indicating potential of using large-scale radiomics for MRI-based diagnosis of CVDs.
Combining Multi-Sequence and Synthetic Images for Improved Segmentation of Late Gadolinium Enhancement Cardiac MRI
Sep 03, 2019



Abstract:Accurate segmentation of the cardiac boundaries in late gadolinium enhancement magnetic resonance images (LGE-MRI) is a fundamental step for accurate quantification of scar tissue. However, while there are many solutions for automatic cardiac segmentation of cine images, the presence of scar tissue can make the correct delineation of the myocardium in LGE-MRI challenging even for human experts. As part of the Multi-Sequence Cardiac MR Segmentation Challenge, we propose a solution for LGE-MRI segmentation based on two components. First, a generative adversarial network is trained for the task of modality-to-modality translation between cine and LGE-MRI sequences to obtain extra synthetic images for both modalities. Second, a deep learning model is trained for segmentation with different combinations of original, augmented and synthetic sequences. Our results based on three magnetic resonance sequences (LGE, bSSFP and T2) from 45 different patients show that the multi-sequence model training integrating synthetic images and data augmentation improves in the segmentation over conventional training with real datasets. In conclusion, the accuracy of the segmentation of LGE-MRI images can be improved by using complementary information provided by non-contrast MRI sequences.
Joint Motion Estimation and Segmentation from Undersampled Cardiac MR Image
Aug 20, 2019


Abstract:Accelerating the acquisition of magnetic resonance imaging (MRI) is a challenging problem, and many works have been proposed to reconstruct images from undersampled k-space data. However, if the main purpose is to extract certain quantitative measures from the images, perfect reconstructions may not always be necessary as long as the images enable the means of extracting the clinically relevant measures. In this paper, we work on jointly predicting cardiac motion estimation and segmentation directly from undersampled data, which are two important steps in quantitatively assessing cardiac function and diagnosing cardiovascular diseases. In particular, a unified model consisting of both motion estimation branch and segmentation branch is learned by optimising the two tasks simultaneously. Additional corresponding fully-sampled images are incorporated into the network as a parallel sub-network to enhance and guide the learning during the training process. Experimental results using cardiac MR images from 220 subjects show that the proposed model is robust to undersampled data and is capable of predicting results that are close to that from fully-sampled ones, while bypassing the usual image reconstruction stage.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge