Athira J Jacob
Fake It Till You Make It: Using Synthetic Data and Domain Knowledge for Improved Text-Based Learning for LGE Detection
Feb 18, 2025



Abstract:Detection of hyperenhancement from cardiac LGE MRI images is a complex task requiring significant clinical expertise. Although deep learning-based models have shown promising results for the task, they require large amounts of data with fine-grained annotations. Clinical reports generated for cardiac MR studies contain rich, clinically relevant information, including the location, extent and etiology of any scars present. Although recently developed CLIP-based training enables pretraining models with image-text pairs, it requires large amounts of data and further finetuning strategies on downstream tasks. In this study, we use various strategies rooted in domain knowledge to train a model for LGE detection solely using text from clinical reports, on a relatively small clinical cohort of 965 patients. We improve performance through the use of synthetic data augmentation, by systematically creating scar images and associated text. In addition, we standardize the orientation of the images in an anatomy-informed way to enable better alignment of spatial and text features. We also use a captioning loss to enable fine-grained supervision and explore the effect of pretraining of the vision encoder on performance. Finally, ablation studies are carried out to elucidate the contributions of each design component to the overall performance of the model.
Towards a vision foundation model for comprehensive assessment of Cardiac MRI
Oct 02, 2024


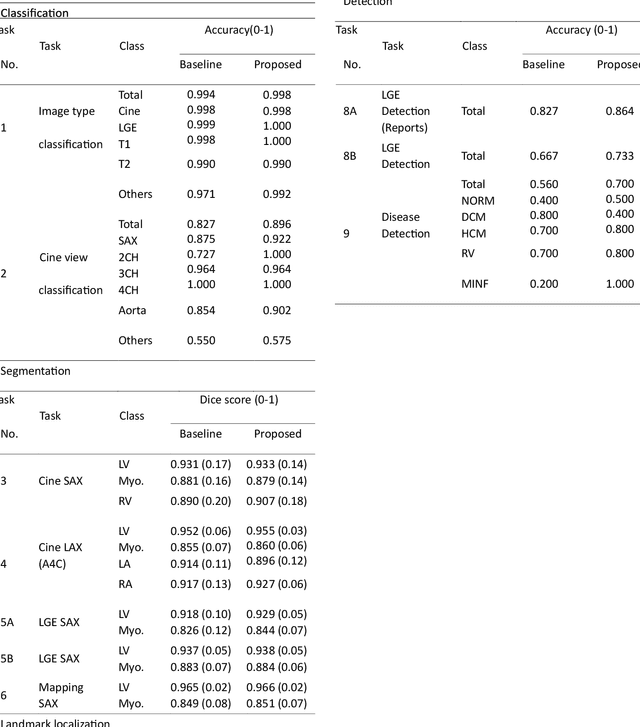
Abstract:Cardiac magnetic resonance imaging (CMR), considered the gold standard for noninvasive cardiac assessment, is a diverse and complex modality requiring a wide variety of image processing tasks for comprehensive assessment of cardiac morphology and function. Advances in deep learning have enabled the development of state-of-the-art (SoTA) models for these tasks. However, model training is challenging due to data and label scarcity, especially in the less common imaging sequences. Moreover, each model is often trained for a specific task, with no connection between related tasks. In this work, we introduce a vision foundation model trained for CMR assessment, that is trained in a self-supervised fashion on 36 million CMR images. We then finetune the model in supervised way for 9 clinical tasks typical to a CMR workflow, across classification, segmentation, landmark localization, and pathology detection. We demonstrate improved accuracy and robustness across all tasks, over a range of available labeled dataset sizes. We also demonstrate improved few-shot learning with fewer labeled samples, a common challenge in medical image analyses. We achieve an out-of-box performance comparable to SoTA for most clinical tasks. The proposed method thus presents a resource-efficient, unified framework for CMR assessment, with the potential to accelerate the development of deep learning-based solutions for image analysis tasks, even with few annotated data available.
DCSM 2.0: Deep Conditional Shape Models for Data Efficient Segmentation
Jun 28, 2024


Abstract:Segmentation is often the first step in many medical image analyses workflows. Deep learning approaches, while giving state-of-the-art accuracies, are data intensive and do not scale well to low data regimes. We introduce Deep Conditional Shape Models 2.0, which uses an edge detector, along with an implicit shape function conditioned on edge maps, to leverage cross-modality shape information. The shape function is trained exclusively on a source domain (contrasted CT) and applied to the target domain of interest (3D echocardiography). We demonstrate data efficiency in the target domain by varying the amounts of training data used in the edge detection stage. We observe that DCSM 2.0 outperforms the baseline at all data levels in terms of Hausdorff distances, and while using 50% or less of the training data in terms of average mesh distance, and at 10% or less of the data with the dice coefficient. The method scales well to low data regimes, with gains of up to 5% in dice coefficient, 2.58 mm in average surface distance and 21.02 mm in Hausdorff distance when using just 2% (22 volumes) of the training data.
AI-based, automated chamber volumetry from gated, non-contrast CT
Oct 25, 2023Abstract:Background: Accurate chamber volumetry from gated, non-contrast cardiac CT (NCCT) scans can be useful for potential screening of heart failure. Objectives: To validate a new, fully automated, AI-based method for cardiac volume and myocardial mass quantification from NCCT scans compared to contrasted CT Angiography (CCTA). Methods: Of a retrospectively collected cohort of 1051 consecutive patients, 420 patients had both NCCT and CCTA scans at mid-diastolic phase, excluding patients with cardiac devices. Ground truth values were obtained from the CCTA scans. Results: The NCCT volume computation shows good agreement with ground truth values. Volume differences [95% CI ] and correlation coefficients were: -9.6 [-45; 26] mL, r = 0.98 for LV Total, -5.4 [-24; 13] mL, r = 0.95 for LA, -8.7 [-45; 28] mL, r = 0.94 for RV, -5.2 [-27; 17] mL, r = 0.92 for RA, -3.2 [-42; 36] mL, r = 0.91 for LV blood pool, and -6.7 [-39; 26] g, r = 0.94 for LV wall mass, respectively. Mean relative volume errors of less than 7% were obtained for all chambers. Conclusions: Fully automated assessment of chamber volumes from NCCT scans is feasible and correlates well with volumes obtained from contrast study.
Deep Conditional Shape Models for 3D cardiac image segmentation
Oct 16, 2023Abstract:Delineation of anatomical structures is often the first step of many medical image analysis workflows. While convolutional neural networks achieve high performance, these do not incorporate anatomical shape information. We introduce a novel segmentation algorithm that uses Deep Conditional Shape models (DCSMs) as a core component. Using deep implicit shape representations, the algorithm learns a modality-agnostic shape model that can generate the signed distance functions for any anatomy of interest. To fit the generated shape to the image, the shape model is conditioned on anatomic landmarks that can be automatically detected or provided by the user. Finally, we add a modality-dependent, lightweight refinement network to capture any fine details not represented by the implicit function. The proposed DCSM framework is evaluated on the problem of cardiac left ventricle (LV) segmentation from multiple 3D modalities (contrast-enhanced CT, non-contrasted CT, 3D echocardiography-3DE). We demonstrate that the automatic DCSM outperforms the baseline for non-contrasted CT without the local refinement, and with the refinement for contrasted CT and 3DE, especially with significant improvement in the Hausdorff distance. The semi-automatic DCSM with user-input landmarks, while only trained on contrasted CT, achieves greater than 92% Dice for all modalities. Both automatic DCSM with refinement and semi-automatic DCSM achieve equivalent or better performance compared to inter-user variability for these modalities.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge