Venkatesh N. Murthy
Do Mice Grok? Glimpses of Hidden Progress During Overtraining in Sensory Cortex
Nov 05, 2024



Abstract:Does learning of task-relevant representations stop when behavior stops changing? Motivated by recent theoretical advances in machine learning and the intuitive observation that human experts continue to learn from practice even after mastery, we hypothesize that task-specific representation learning can continue, even when behavior plateaus. In a novel reanalysis of recently published neural data, we find evidence for such learning in posterior piriform cortex of mice following continued training on a task, long after behavior saturates at near-ceiling performance ("overtraining"). This learning is marked by an increase in decoding accuracy from piriform neural populations and improved performance on held-out generalization tests. We demonstrate that class representations in cortex continue to separate during overtraining, so that examples that were incorrectly classified at the beginning of overtraining can abruptly be correctly classified later on, despite no changes in behavior during that time. We hypothesize this hidden yet rich learning takes the form of approximate margin maximization; we validate this and other predictions in the neural data, as well as build and interpret a simple synthetic model that recapitulates these phenomena. We conclude by showing how this model of late-time feature learning implies an explanation for the empirical puzzle of overtraining reversal in animal learning, where task-specific representations are more robust to particular task changes because the learned features can be reused.
Self-Supervised Learning for Interventional Image Analytics: Towards Robust Device Trackers
May 02, 2024Abstract:An accurate detection and tracking of devices such as guiding catheters in live X-ray image acquisitions is an essential prerequisite for endovascular cardiac interventions. This information is leveraged for procedural guidance, e.g., directing stent placements. To ensure procedural safety and efficacy, there is a need for high robustness no failures during tracking. To achieve that, one needs to efficiently tackle challenges, such as: device obscuration by contrast agent or other external devices or wires, changes in field-of-view or acquisition angle, as well as the continuous movement due to cardiac and respiratory motion. To overcome the aforementioned challenges, we propose a novel approach to learn spatio-temporal features from a very large data cohort of over 16 million interventional X-ray frames using self-supervision for image sequence data. Our approach is based on a masked image modeling technique that leverages frame interpolation based reconstruction to learn fine inter-frame temporal correspondences. The features encoded in the resulting model are fine-tuned downstream. Our approach achieves state-of-the-art performance and in particular robustness compared to ultra optimized reference solutions (that use multi-stage feature fusion, multi-task and flow regularization). The experiments show that our method achieves 66.31% reduction in maximum tracking error against reference solutions (23.20% when flow regularization is used); achieving a success score of 97.95% at a 3x faster inference speed of 42 frames-per-second (on GPU). The results encourage the use of our approach in various other tasks within interventional image analytics that require effective understanding of spatio-temporal semantics.
Markerless tracking of user-defined features with deep learning
Apr 09, 2018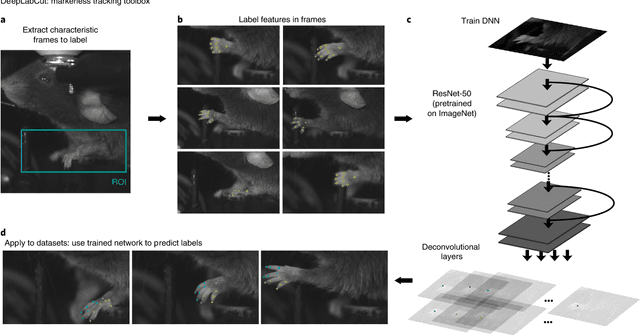
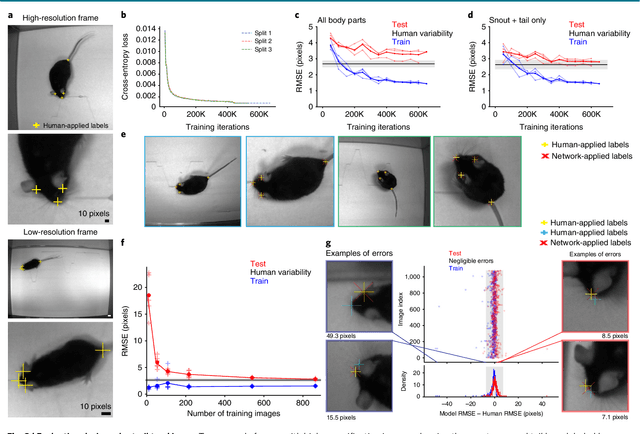
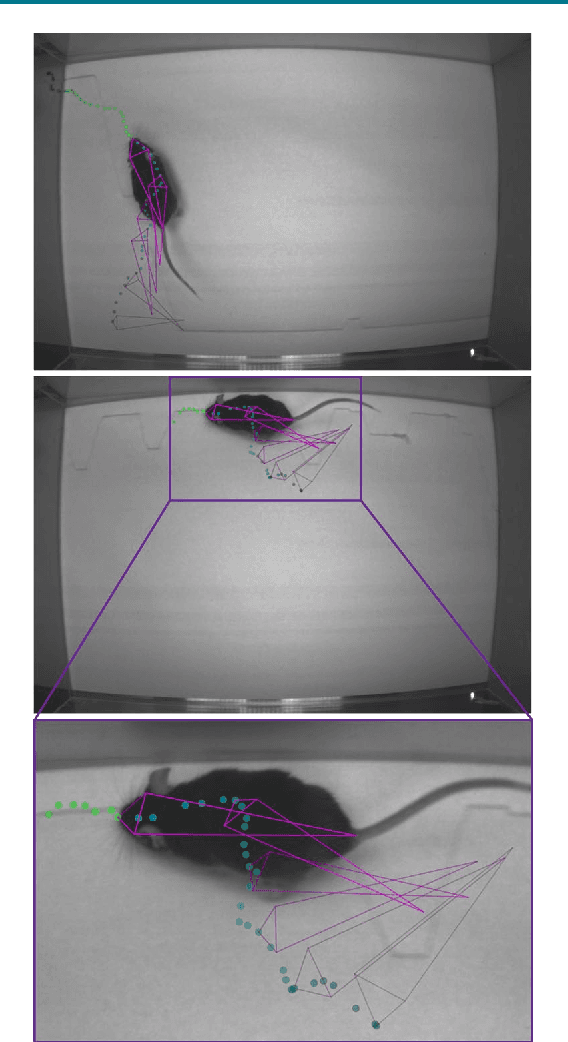
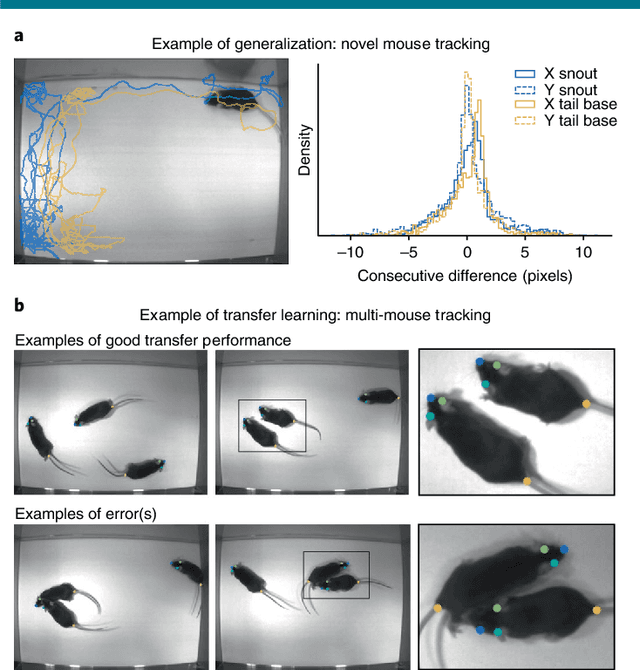
Abstract:Quantifying behavior is crucial for many applications in neuroscience. Videography provides easy methods for the observation and recording of animal behavior in diverse settings, yet extracting particular aspects of a behavior for further analysis can be highly time consuming. In motor control studies, humans or other animals are often marked with reflective markers to assist with computer-based tracking, yet markers are intrusive (especially for smaller animals), and the number and location of the markers must be determined a priori. Here, we present a highly efficient method for markerless tracking based on transfer learning with deep neural networks that achieves excellent results with minimal training data. We demonstrate the versatility of this framework by tracking various body parts in a broad collection of experimental settings: mice odor trail-tracking, egg-laying behavior in drosophila, and mouse hand articulation in a skilled forelimb task. For example, during the skilled reaching behavior, individual joints can be automatically tracked (and a confidence score is reported). Remarkably, even when a small number of frames are labeled ($\approx 200$), the algorithm achieves excellent tracking performance on test frames that is comparable to human accuracy.
* Videos at http://www.mousemotorlab.org/deeplabcut
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge