Guillaume Chabin
COSST: Multi-organ Segmentation with Partially Labeled Datasets Using Comprehensive Supervisions and Self-training
Apr 28, 2023Abstract:Deep learning models have demonstrated remarkable success in multi-organ segmentation but typically require large-scale datasets with all organs of interest annotated. However, medical image datasets are often low in sample size and only partially labeled, i.e., only a subset of organs are annotated. Therefore, it is crucial to investigate how to learn a unified model on the available partially labeled datasets to leverage their synergistic potential. In this paper, we empirically and systematically study the partial-label segmentation with in-depth analyses on the existing approaches and identify three distinct types of supervision signals, including two signals derived from ground truth and one from pseudo label. We propose a novel training framework termed COSST, which effectively and efficiently integrates comprehensive supervision signals with self-training. Concretely, we first train an initial unified model using two ground truth-based signals and then iteratively incorporate the pseudo label signal to the initial model using self-training. To mitigate performance degradation caused by unreliable pseudo labels, we assess the reliability of pseudo labels via outlier detection in latent space and exclude the most unreliable pseudo labels from each self-training iteration. Extensive experiments are conducted on six CT datasets for three partial-label segmentation tasks. Experimental results show that our proposed COSST achieves significant improvement over the baseline method, i.e., individual networks trained on each partially labeled dataset. Compared to the state-of-the-art partial-label segmentation methods, COSST demonstrates consistent superior performance on various segmentation tasks and with different training data size.
Automated detection and quantification of COVID-19 airspace disease on chest radiographs: A novel approach achieving radiologist-level performance using a CNN trained on digital reconstructed radiographs (DRRs) from CT-based ground-truth
Aug 13, 2020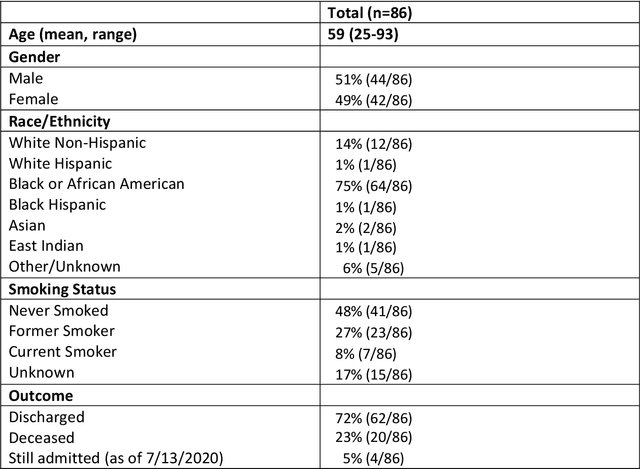
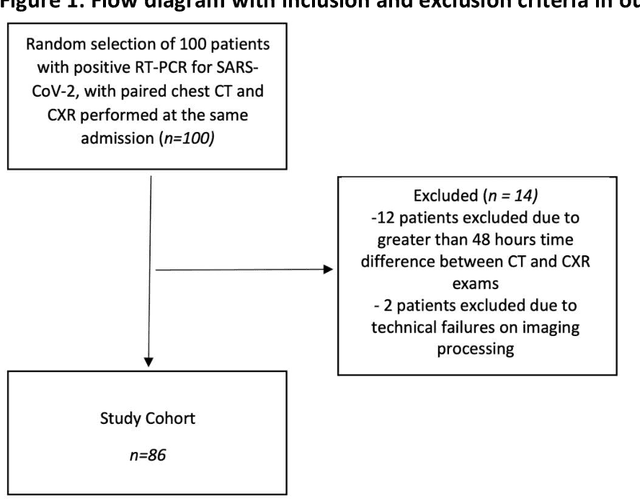
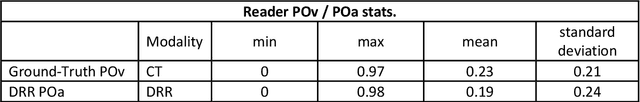
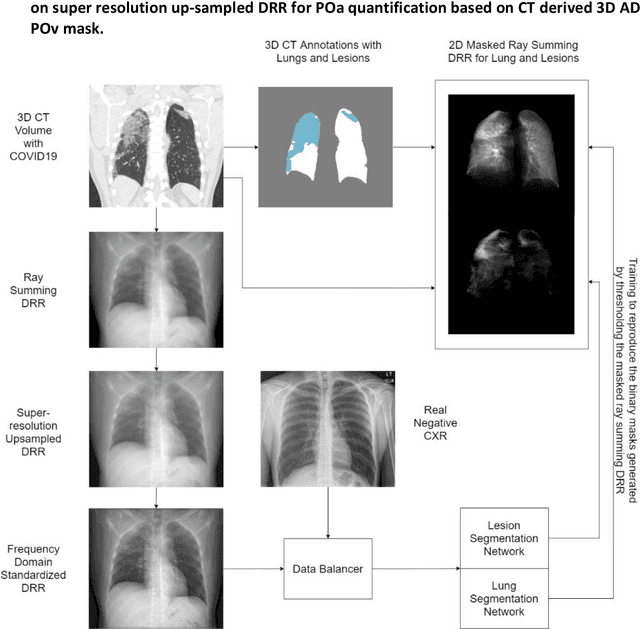
Abstract:Purpose: To leverage volumetric quantification of airspace disease (AD) derived from a superior modality (CT) serving as ground truth, projected onto digitally reconstructed radiographs (DRRs) to: 1) train a convolutional neural network to quantify airspace disease on paired CXRs; and 2) compare the DRR-trained CNN to expert human readers in the CXR evaluation of patients with confirmed COVID-19. Materials and Methods: We retrospectively selected a cohort of 86 COVID-19 patients (with positive RT-PCR), from March-May 2020 at a tertiary hospital in the northeastern USA, who underwent chest CT and CXR within 48 hrs. The ground truth volumetric percentage of COVID-19 related AD (POv) was established by manual AD segmentation on CT. The resulting 3D masks were projected into 2D anterior-posterior digitally reconstructed radiographs (DRR) to compute area-based AD percentage (POa). A convolutional neural network (CNN) was trained with DRR images generated from a larger-scale CT dataset of COVID-19 and non-COVID-19 patients, automatically segmenting lungs, AD and quantifying POa on CXR. CNN POa results were compared to POa quantified on CXR by two expert readers and to the POv ground-truth, by computing correlations and mean absolute errors. Results: Bootstrap mean absolute error (MAE) and correlations between POa and POv were 11.98% [11.05%-12.47%] and 0.77 [0.70-0.82] for average of expert readers, and 9.56%-9.78% [8.83%-10.22%] and 0.78-0.81 [0.73-0.85] for the CNN, respectively. Conclusion: Our CNN trained with DRR using CT-derived airspace quantification achieved expert radiologist level of accuracy in the quantification of airspace disease on CXR, in patients with positive RT-PCR for COVID-19.
Machine Learning Automatically Detects COVID-19 using Chest CTs in a Large Multicenter Cohort
Jun 11, 2020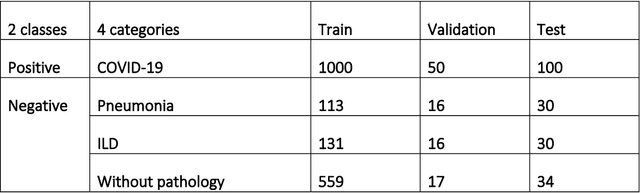
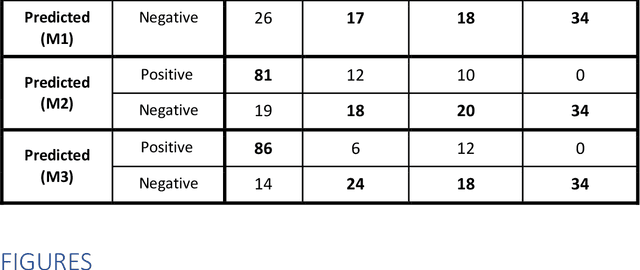
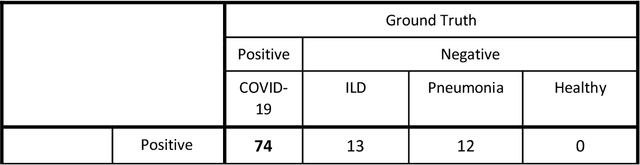
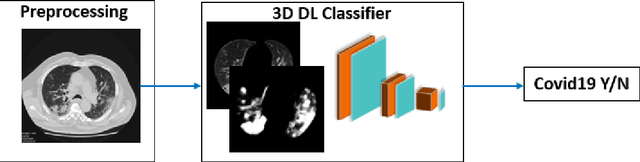
Abstract:Purpose: To investigate if AI-based classifiers can distinguish COVID-19 from other pulmonary diseases and normal groups, using chest CT images. To study the interpretability of discriminative features for COVID19 detection. Materials and Methods: Our database consists of 2096 CT exams that include CTs from 1150 COVID-19 patients. Training was performed on 1000 COVID-19, 131 ILD, 113 other pneumonias, 559 normal CTs, and testing on 100 COVID-19, 30 ILD, 30 other pneumonias, and 34 normal CTs. A metric-based approach for classification of COVID-19 used interpretable features, relying on logistic regression and random forests. A deep learning-based classifier differentiated COVID-19 based on 3D features extracted directly from CT intensities and from the probability distribution of airspace opacities. Results: Most discriminative features of COVID-19 are percentage of airspace opacity, ground glass opacities, consolidations, and peripheral and basal opacities, which coincide with the typical characterization of COVID-19 in the literature. Unsupervised hierarchical clustering compares the distribution of these features across COVID-19 and control cohorts. The metrics-based classifier achieved AUC, sensitivity, and specificity of respectively 0.85, 0.81, and 0.77. The DL-based classifier achieved AUC, sensitivity, and specificity of respectively 0.90, 0.86, and 0.81. Most of ambiguity comes from non-COVID-19 pneumonia with manifestations that overlap with COVID-19, as well as COVID-19 cases in early stages. Conclusion: A new method discriminates COVID-19 from other types of pneumonia, ILD, and normal, using quantitative patterns from chest CT. Our models balance interpretability of results and classification performance, and therefore may be useful to expedite and improve diagnosis of COVID-19.
3D Tomographic Pattern Synthesis for Enhancing the Quantification of COVID-19
May 05, 2020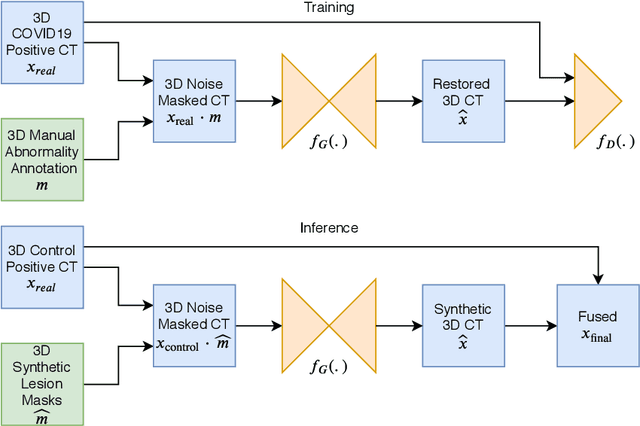
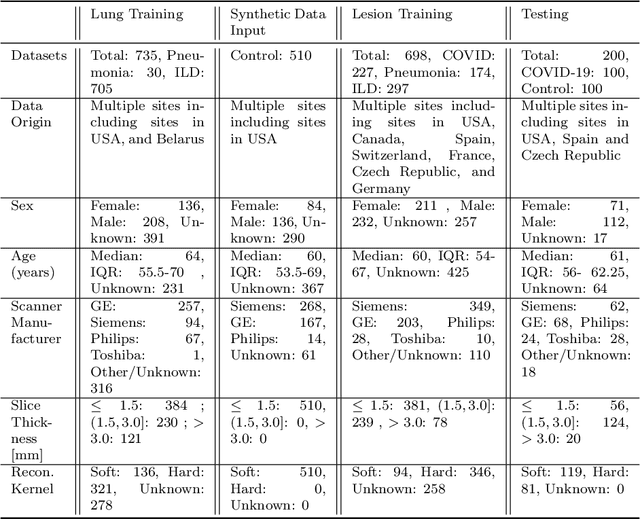
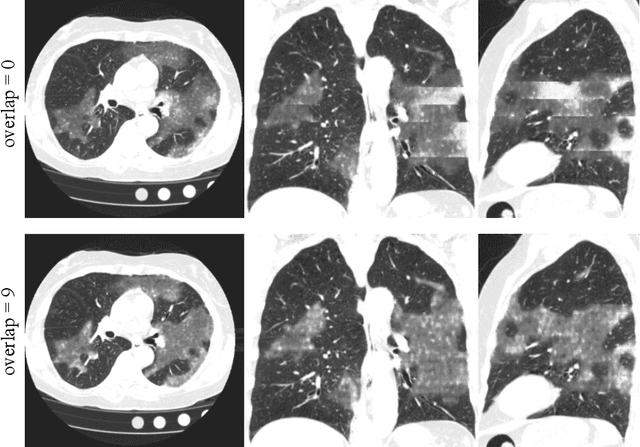
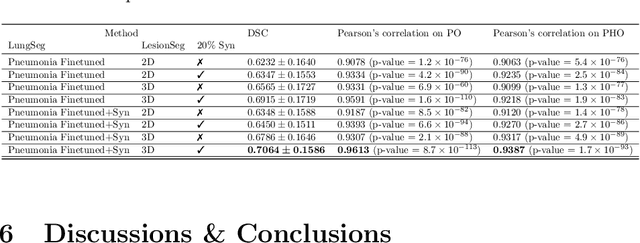
Abstract:The Coronavirus Disease (COVID-19) has affected 1.8 million people and resulted in more than 110,000 deaths as of April 12, 2020. Several studies have shown that tomographic patterns seen on chest Computed Tomography (CT), such as ground-glass opacities, consolidations, and crazy paving pattern, are correlated with the disease severity and progression. CT imaging can thus emerge as an important modality for the management of COVID-19 patients. AI-based solutions can be used to support CT based quantitative reporting and make reading efficient and reproducible if quantitative biomarkers, such as the Percentage of Opacity (PO), can be automatically computed. However, COVID-19 has posed unique challenges to the development of AI, specifically concerning the availability of appropriate image data and annotations at scale. In this paper, we propose to use synthetic datasets to augment an existing COVID-19 database to tackle these challenges. We train a Generative Adversarial Network (GAN) to inpaint COVID-19 related tomographic patterns on chest CTs from patients without infectious diseases. Additionally, we leverage location priors derived from manually labeled COVID-19 chest CTs patients to generate appropriate abnormality distributions. Synthetic data are used to improve both lung segmentation and segmentation of COVID-19 patterns by adding 20% of synthetic data to the real COVID-19 training data. We collected 2143 chest CTs, containing 327 COVID-19 positive cases, acquired from 12 sites across 7 countries. By testing on 100 COVID-19 positive and 100 control cases, we show that synthetic data can help improve both lung segmentation (+6.02% lesion inclusion rate) and abnormality segmentation (+2.78% dice coefficient), leading to an overall more accurate PO computation (+2.82% Pearson coefficient).
Quantification of Tomographic Patterns associated with COVID-19 from Chest CT
Apr 28, 2020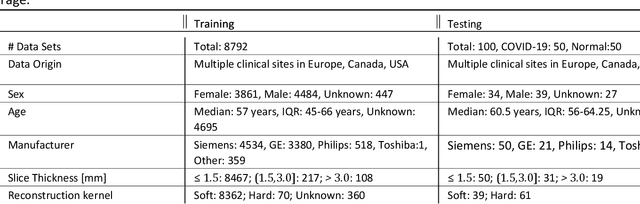
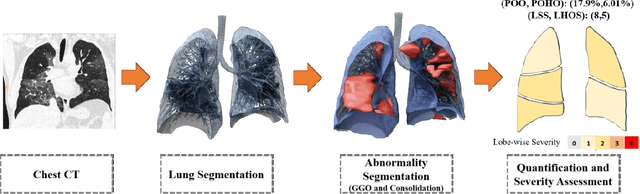
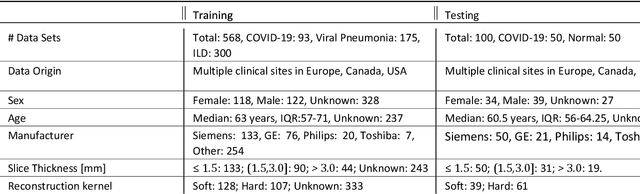
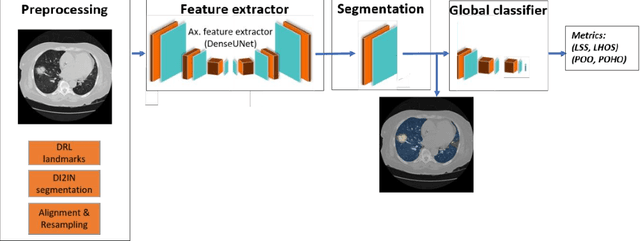
Abstract:Purpose: To present a method that automatically detects and quantifies abnormal tomographic patterns commonly present in COVID-19, namely Ground Glass Opacities (GGO) and consolidations. Given that high opacity abnormalities (i.e., consolidations) were shown to correlate with severe disease, the paper introduces two combined severity measures (Percentage of Opacity, Percentage of High Opacity) and (Lung Severity Score, Lung High Opacity Score). They quantify the extent of overall COVID-19 abnormalities and the presence of high opacity abnormalities, global and lobe-wise, respectively, being computed based on 3D segmentations of lesions, lungs, and lobes. Materials and Methods: The proposed method takes as input a non-contrasted Chest CT and segments the lesions, lungs, and lobes in 3D. It outputs two combined measures of the severity of lung/lobe involvement, quantifying both the extent of COVID-19 abnormalities and presence of high opacities, based on deep learning and deep reinforcement learning. The first measure (POO, POHO) is global, while the second (LSS, LHOS) is lobe-wise. Evaluation is reported on CTs of 100 subjects (50 COVID-19 confirmed and 50 controls) from institutions from Canada, Europe and US. Ground truth is established by manual annotations of lesions, lungs, and lobes. Results: Pearson Correlation Coefficient between method prediction and ground truth is 0.97 (POO), 0.98 (POHO), 0.96 (LSS), 0.96 (LHOS). Automated processing time to compute the severity scores is 10 seconds/case vs 30 mins needed for manual annotations. Conclusion: A new method identifies regions of abnormalities seen in COVID-19 non-contrasted Chest CT and computes (POO, POHO) and (LSS, LHOS) severity scores.
Class-Aware Adversarial Lung Nodule Synthesis in CT Images
Dec 28, 2018



Abstract:Though large-scale datasets are essential for training deep learning systems, it is expensive to scale up the collection of medical imaging datasets. Synthesizing the objects of interests, such as lung nodules, in medical images based on the distribution of annotated datasets can be helpful for improving the supervised learning tasks, especially when the datasets are limited by size and class balance. In this paper, we propose the class-aware adversarial synthesis framework to synthesize lung nodules in CT images. The framework is built with a coarse-to-fine patch in-painter (generator) and two class-aware discriminators. By conditioning on the random latent variables and the target nodule labels, the trained networks are able to generate diverse nodules given the same context. By evaluating on the public LIDC-IDRI dataset, we demonstrate an example application of the proposed framework for improving the accuracy of the lung nodule malignancy estimation as a binary classification problem, which is important in the lung screening scenario. We show that combining the real image patches and the synthetic lung nodules in the training set can improve the mean AUC classification score across different network architectures by 2%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge