Brian Teixeira
Automated CT Lung Cancer Screening Workflow using 3D Camera
Sep 27, 2023Abstract:Despite recent developments in CT planning that enabled automation in patient positioning, time-consuming scout scans are still needed to compute dose profile and ensure the patient is properly positioned. In this paper, we present a novel method which eliminates the need for scout scans in CT lung cancer screening by estimating patient scan range, isocenter, and Water Equivalent Diameter (WED) from 3D camera images. We achieve this task by training an implicit generative model on over 60,000 CT scans and introduce a novel approach for updating the prediction using real-time scan data. We demonstrate the effectiveness of our method on a testing set of 110 pairs of depth data and CT scan, resulting in an average error of 5mm in estimating the isocenter, 13mm in determining the scan range, 10mm and 16mm in estimating the AP and lateral WED respectively. The relative WED error of our method is 4%, which is well within the International Electrotechnical Commission (IEC) acceptance criteria of 10%.
View Invariant Human Body Detection and Pose Estimation from Multiple Depth Sensors
May 08, 2020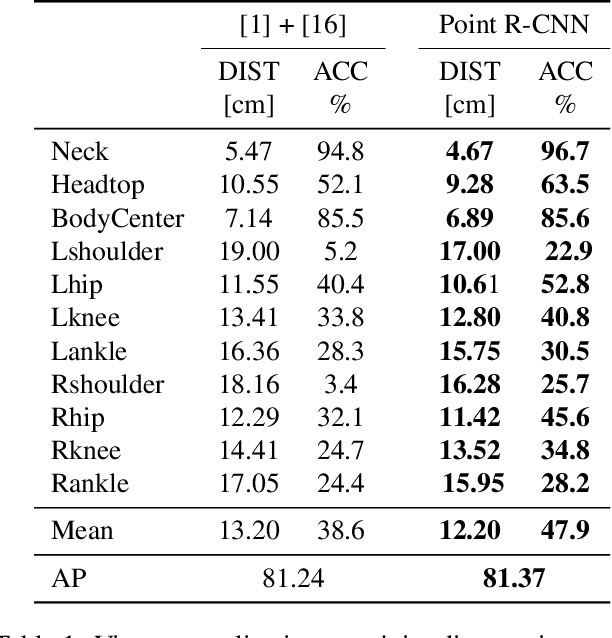
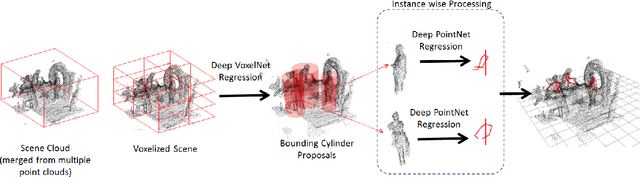
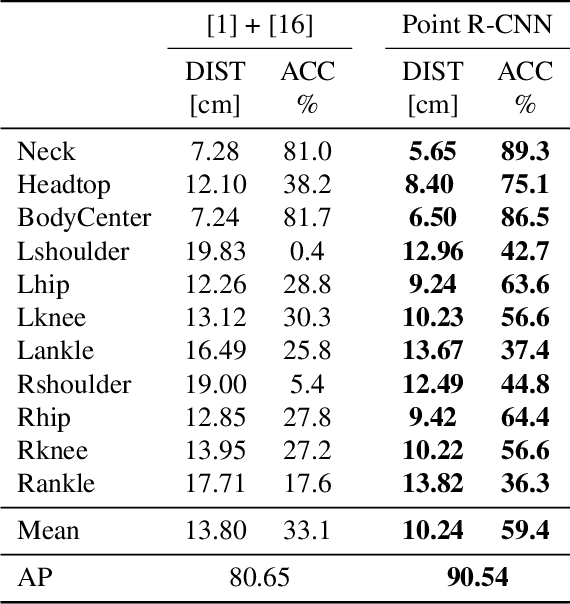
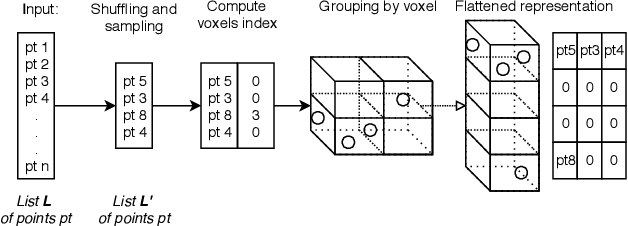
Abstract:Point cloud based methods have produced promising results in areas such as 3D object detection in autonomous driving. However, most of the recent point cloud work focuses on single depth sensor data, whereas less work has been done on indoor monitoring applications, such as operation room monitoring in hospitals or indoor surveillance. In these scenarios multiple cameras are often used to tackle occlusion problems. We propose an end-to-end multi-person 3D pose estimation network, Point R-CNN, using multiple point cloud sources. We conduct extensive experiments to simulate challenging real world cases, such as individual camera failures, various target appearances, and complex cluttered scenes with the CMU panoptic dataset and the MVOR operation room dataset. Unlike most of the previous methods that attempt to use multiple sensor information by building complex fusion models, which often lead to poor generalization, we take advantage of the efficiency of concatenating point clouds to fuse the information at the input level. In the meantime, we show our end-to-end network greatly outperforms cascaded state-of-the-art models.
3D Tomographic Pattern Synthesis for Enhancing the Quantification of COVID-19
May 05, 2020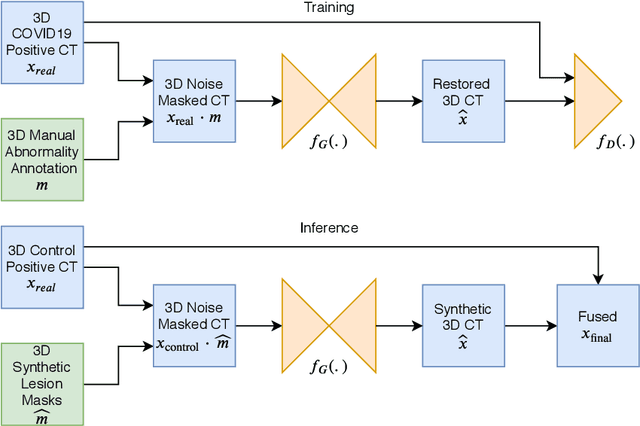
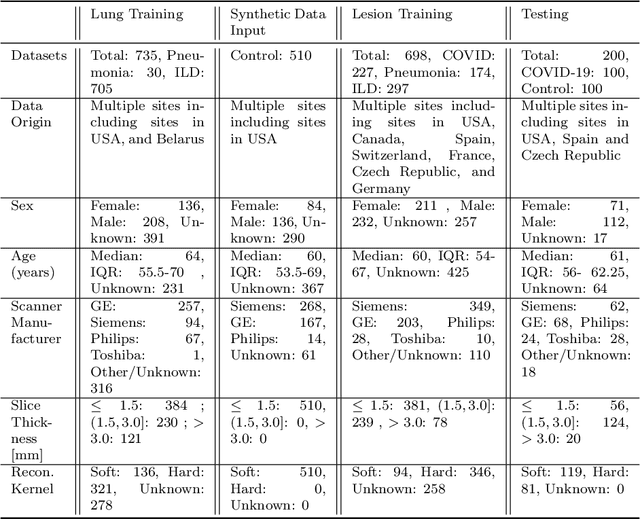
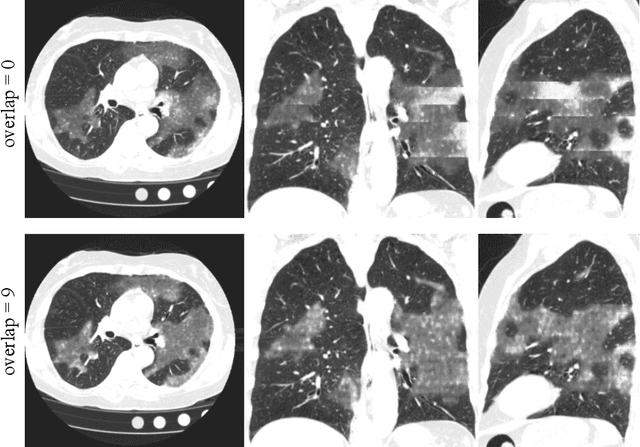
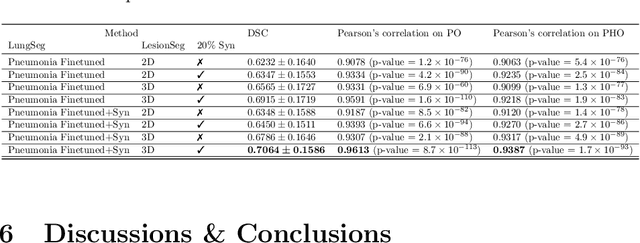
Abstract:The Coronavirus Disease (COVID-19) has affected 1.8 million people and resulted in more than 110,000 deaths as of April 12, 2020. Several studies have shown that tomographic patterns seen on chest Computed Tomography (CT), such as ground-glass opacities, consolidations, and crazy paving pattern, are correlated with the disease severity and progression. CT imaging can thus emerge as an important modality for the management of COVID-19 patients. AI-based solutions can be used to support CT based quantitative reporting and make reading efficient and reproducible if quantitative biomarkers, such as the Percentage of Opacity (PO), can be automatically computed. However, COVID-19 has posed unique challenges to the development of AI, specifically concerning the availability of appropriate image data and annotations at scale. In this paper, we propose to use synthetic datasets to augment an existing COVID-19 database to tackle these challenges. We train a Generative Adversarial Network (GAN) to inpaint COVID-19 related tomographic patterns on chest CTs from patients without infectious diseases. Additionally, we leverage location priors derived from manually labeled COVID-19 chest CTs patients to generate appropriate abnormality distributions. Synthetic data are used to improve both lung segmentation and segmentation of COVID-19 patterns by adding 20% of synthetic data to the real COVID-19 training data. We collected 2143 chest CTs, containing 327 COVID-19 positive cases, acquired from 12 sites across 7 countries. By testing on 100 COVID-19 positive and 100 control cases, we show that synthetic data can help improve both lung segmentation (+6.02% lesion inclusion rate) and abnormality segmentation (+2.78% dice coefficient), leading to an overall more accurate PO computation (+2.82% Pearson coefficient).
Adaloss: Adaptive Loss Function for Landmark Localization
Aug 02, 2019



Abstract:Landmark localization is a challenging problem in computer vision with a multitude of applications. Recent deep learning based methods have shown improved results by regressing likelihood maps instead of regressing the coordinates directly. However, setting the precision of these regression targets during the training is a cumbersome process since it creates a trade-off between trainability vs localization accuracy. Using precise targets introduces a significant sampling bias and hence makes the training more difficult, whereas using imprecise targets results in inaccurate landmark detectors. In this paper, we introduce "Adaloss", an objective function that adapts itself during the training by updating the target precision based on the training statistics. This approach does not require setting problem-specific parameters and shows improved stability in training and better localization accuracy during inference. We demonstrate the effectiveness of our proposed method in three different applications of landmark localization: 1) the challenging task of precisely detecting catheter tips in medical X-ray images, 2) localizing surgical instruments in endoscopic images, and 3) localizing facial features on in-the-wild images where we show state-of-the-art results on the 300-W benchmark dataset.
3D Organ Shape Reconstruction from Topogram Images
Mar 29, 2019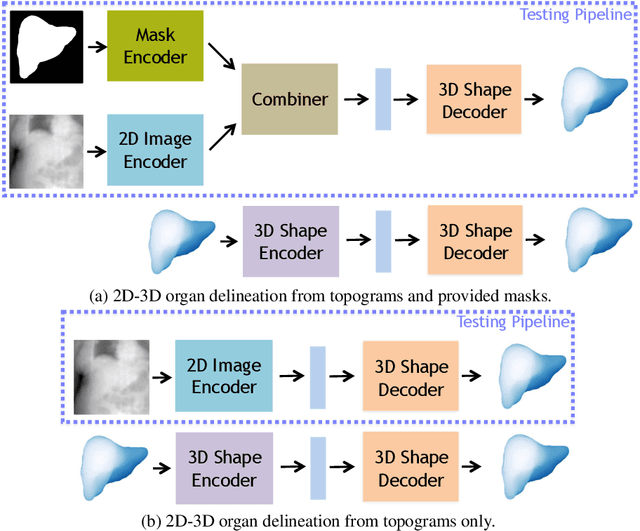

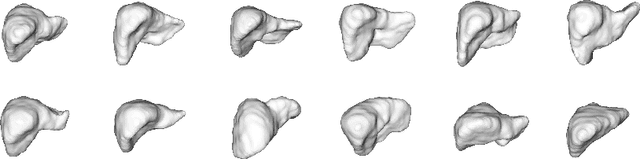

Abstract:Automatic delineation and measurement of main organs such as liver is one of the critical steps for assessment of hepatic diseases, planning and postoperative or treatment follow-up. However, addressing this problem typically requires performing computed tomography (CT) scanning and complicated postprocessing of the resulting scans using slice-by-slice techniques. In this paper, we show that 3D organ shape can be automatically predicted directly from topogram images, which are easier to acquire and have limited exposure to radiation during acquisition, compared to CT scans. We evaluate our approach on the challenging task of predicting liver shape using a generative model. We also demonstrate that our method can be combined with user annotations, such as a 2D mask, for improved prediction accuracy. We show compelling results on 3D liver shape reconstruction and volume estimation on 2129 CT scans.
Structure-Aware Shape Synthesis
Aug 04, 2018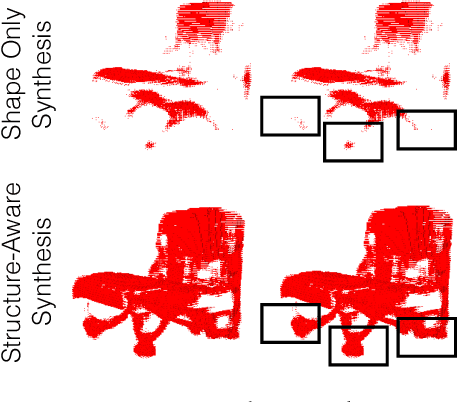

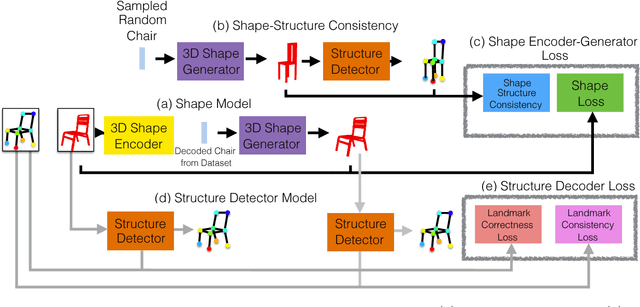
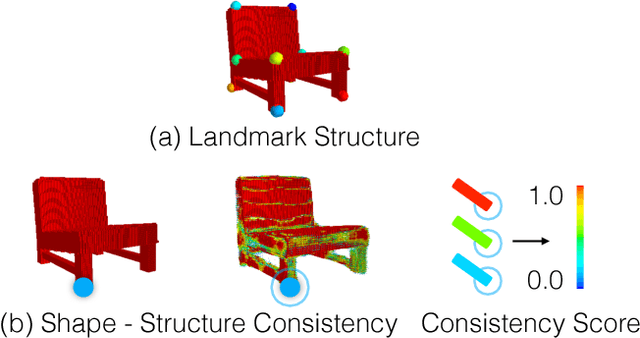
Abstract:We propose a new procedure to guide training of a data-driven shape generative model using a structure-aware loss function. Complex 3D shapes often can be summarized using a coarsely defined structure which is consistent and robust across variety of observations. However, existing synthesis techniques do not account for structure during training, and thus often generate implausible and structurally unrealistic shapes. During training, we enforce structural constraints in order to enforce consistency and structure across the entire manifold. We propose a novel methodology for training 3D generative models that incorporates structural information into an end-to-end training pipeline.
Generating Synthetic X-ray Images of a Person from the Surface Geometry
May 14, 2018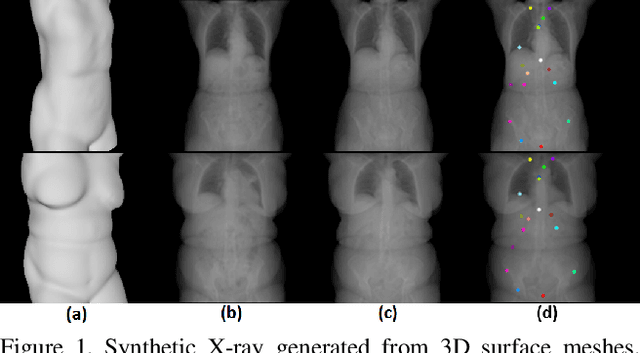
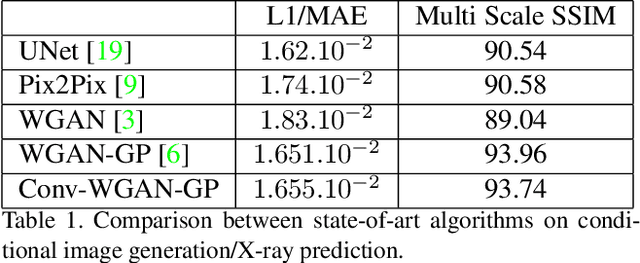
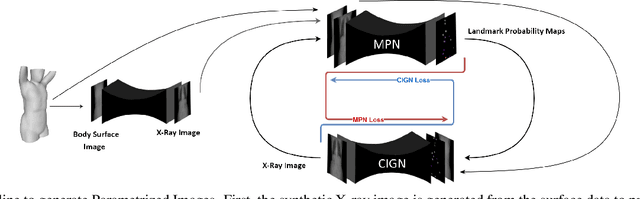
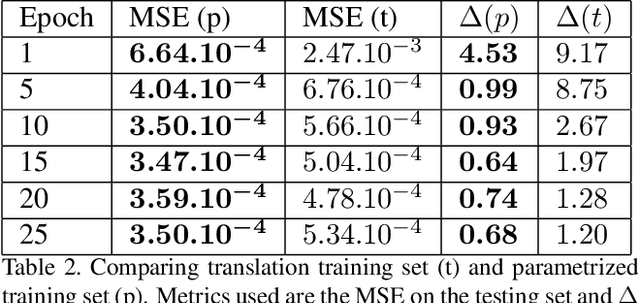
Abstract:We present a novel framework that learns to predict human anatomy from body surface. Specifically, our approach generates a synthetic X-ray image of a person only from the person's surface geometry. Furthermore, the synthetic X-ray image is parametrized and can be manipulated by adjusting a set of body markers which are also generated during the X-ray image prediction. With the proposed framework, multiple synthetic X-ray images can easily be generated by varying surface geometry. By perturbing the parameters, several additional synthetic X-ray images can be generated from the same surface geometry. As a result, our approach offers a potential to overcome the training data barrier in the medical domain. This capability is achieved by learning a pair of networks - one learns to generate the full image from the partial image and a set of parameters, and the other learns to estimate the parameters given the full image. During training, the two networks are trained iteratively such that they would converge to a solution where the predicted parameters and the full image are consistent with each other. In addition to medical data enrichment, our framework can also be used for image completion as well as anomaly detection.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge