Shikha Chaganti
Machine Learning Automatically Detects COVID-19 using Chest CTs in a Large Multicenter Cohort
Jun 11, 2020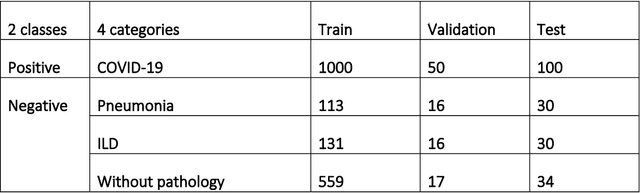
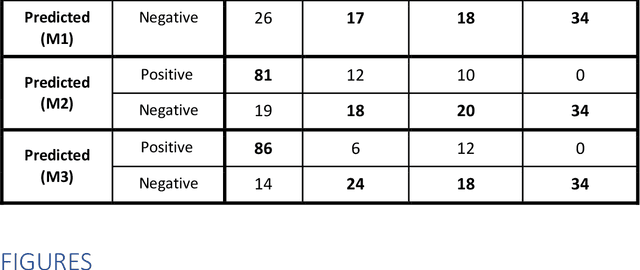
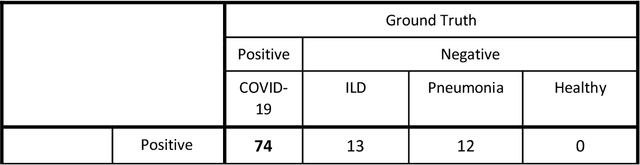
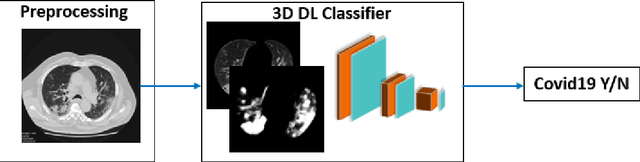
Abstract:Purpose: To investigate if AI-based classifiers can distinguish COVID-19 from other pulmonary diseases and normal groups, using chest CT images. To study the interpretability of discriminative features for COVID19 detection. Materials and Methods: Our database consists of 2096 CT exams that include CTs from 1150 COVID-19 patients. Training was performed on 1000 COVID-19, 131 ILD, 113 other pneumonias, 559 normal CTs, and testing on 100 COVID-19, 30 ILD, 30 other pneumonias, and 34 normal CTs. A metric-based approach for classification of COVID-19 used interpretable features, relying on logistic regression and random forests. A deep learning-based classifier differentiated COVID-19 based on 3D features extracted directly from CT intensities and from the probability distribution of airspace opacities. Results: Most discriminative features of COVID-19 are percentage of airspace opacity, ground glass opacities, consolidations, and peripheral and basal opacities, which coincide with the typical characterization of COVID-19 in the literature. Unsupervised hierarchical clustering compares the distribution of these features across COVID-19 and control cohorts. The metrics-based classifier achieved AUC, sensitivity, and specificity of respectively 0.85, 0.81, and 0.77. The DL-based classifier achieved AUC, sensitivity, and specificity of respectively 0.90, 0.86, and 0.81. Most of ambiguity comes from non-COVID-19 pneumonia with manifestations that overlap with COVID-19, as well as COVID-19 cases in early stages. Conclusion: A new method discriminates COVID-19 from other types of pneumonia, ILD, and normal, using quantitative patterns from chest CT. Our models balance interpretability of results and classification performance, and therefore may be useful to expedite and improve diagnosis of COVID-19.
3D Tomographic Pattern Synthesis for Enhancing the Quantification of COVID-19
May 05, 2020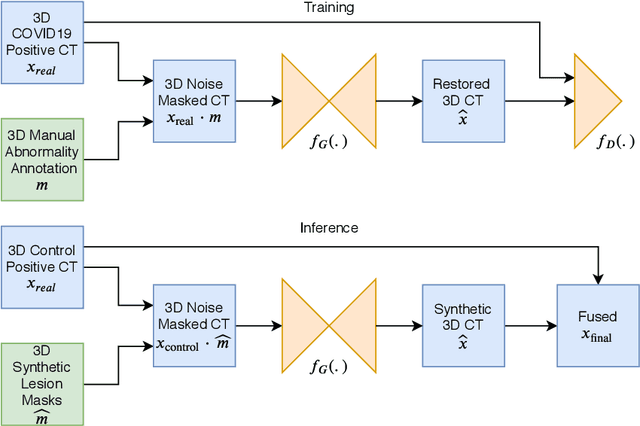
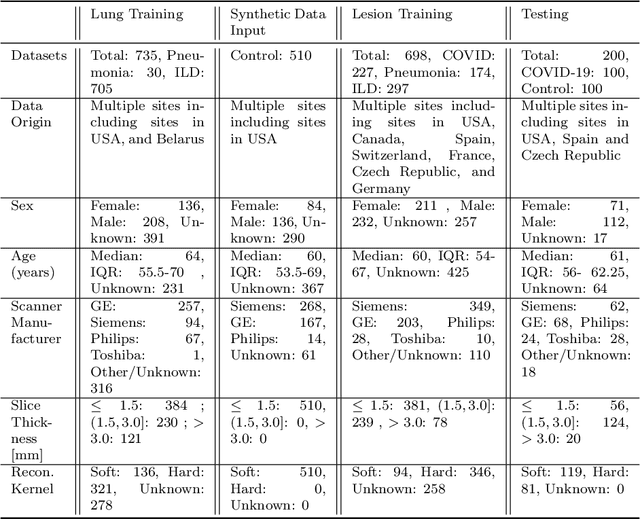
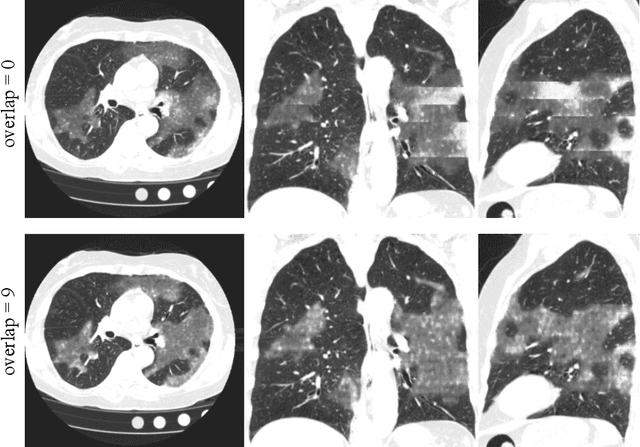
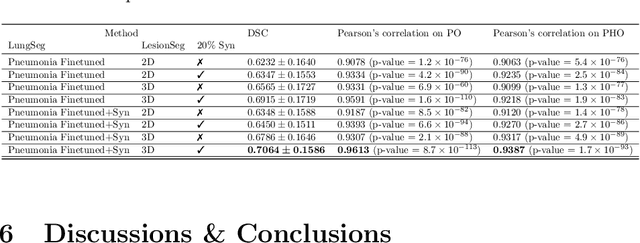
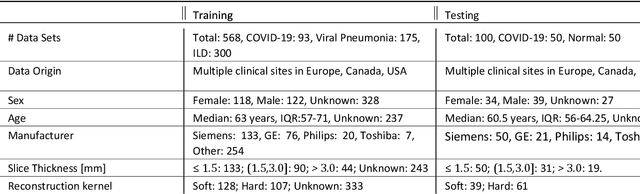
Abstract:The Coronavirus Disease (COVID-19) has affected 1.8 million people and resulted in more than 110,000 deaths as of April 12, 2020. Several studies have shown that tomographic patterns seen on chest Computed Tomography (CT), such as ground-glass opacities, consolidations, and crazy paving pattern, are correlated with the disease severity and progression. CT imaging can thus emerge as an important modality for the management of COVID-19 patients. AI-based solutions can be used to support CT based quantitative reporting and make reading efficient and reproducible if quantitative biomarkers, such as the Percentage of Opacity (PO), can be automatically computed. However, COVID-19 has posed unique challenges to the development of AI, specifically concerning the availability of appropriate image data and annotations at scale. In this paper, we propose to use synthetic datasets to augment an existing COVID-19 database to tackle these challenges. We train a Generative Adversarial Network (GAN) to inpaint COVID-19 related tomographic patterns on chest CTs from patients without infectious diseases. Additionally, we leverage location priors derived from manually labeled COVID-19 chest CTs patients to generate appropriate abnormality distributions. Synthetic data are used to improve both lung segmentation and segmentation of COVID-19 patterns by adding 20% of synthetic data to the real COVID-19 training data. We collected 2143 chest CTs, containing 327 COVID-19 positive cases, acquired from 12 sites across 7 countries. By testing on 100 COVID-19 positive and 100 control cases, we show that synthetic data can help improve both lung segmentation (+6.02% lesion inclusion rate) and abnormality segmentation (+2.78% dice coefficient), leading to an overall more accurate PO computation (+2.82% Pearson coefficient).
Quantification of Tomographic Patterns associated with COVID-19 from Chest CT
Apr 28, 2020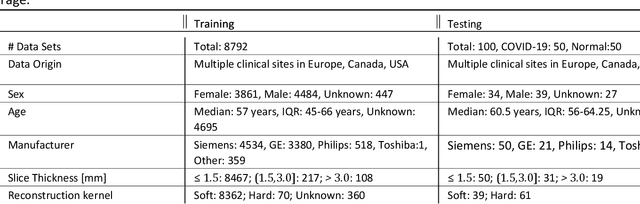
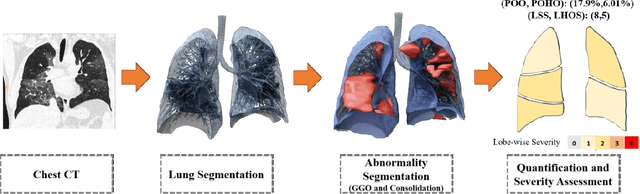
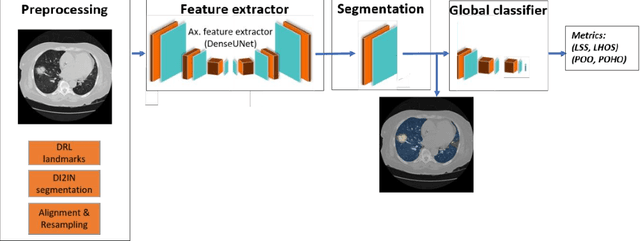
Abstract:Purpose: To present a method that automatically detects and quantifies abnormal tomographic patterns commonly present in COVID-19, namely Ground Glass Opacities (GGO) and consolidations. Given that high opacity abnormalities (i.e., consolidations) were shown to correlate with severe disease, the paper introduces two combined severity measures (Percentage of Opacity, Percentage of High Opacity) and (Lung Severity Score, Lung High Opacity Score). They quantify the extent of overall COVID-19 abnormalities and the presence of high opacity abnormalities, global and lobe-wise, respectively, being computed based on 3D segmentations of lesions, lungs, and lobes. Materials and Methods: The proposed method takes as input a non-contrasted Chest CT and segments the lesions, lungs, and lobes in 3D. It outputs two combined measures of the severity of lung/lobe involvement, quantifying both the extent of COVID-19 abnormalities and presence of high opacities, based on deep learning and deep reinforcement learning. The first measure (POO, POHO) is global, while the second (LSS, LHOS) is lobe-wise. Evaluation is reported on CTs of 100 subjects (50 COVID-19 confirmed and 50 controls) from institutions from Canada, Europe and US. Ground truth is established by manual annotations of lesions, lungs, and lobes. Results: Pearson Correlation Coefficient between method prediction and ground truth is 0.97 (POO), 0.98 (POHO), 0.96 (LSS), 0.96 (LHOS). Automated processing time to compute the severity scores is 10 seconds/case vs 30 mins needed for manual annotations. Conclusion: A new method identifies regions of abnormalities seen in COVID-19 non-contrasted Chest CT and computes (POO, POHO) and (LSS, LHOS) severity scores.
Graph Attention Network based Pruning for Reconstructing 3D Liver Vessel Morphology from Contrasted CT Images
Mar 18, 2020



Abstract:With the injection of contrast material into blood vessels, multi-phase contrasted CT images can enhance the visibility of vessel networks in the human body. Reconstructing the 3D geometric morphology of liver vessels from the contrasted CT images can enable multiple liver preoperative surgical planning applications. Automatic reconstruction of liver vessel morphology remains a challenging problem due to the morphological complexity of liver vessels and the inconsistent vessel intensities among different multi-phase contrasted CT images. On the other side, high integrity is required for the 3D reconstruction to avoid decision making biases. In this paper, we propose a framework for liver vessel morphology reconstruction using both a fully convolutional neural network and a graph attention network. A fully convolutional neural network is first trained to produce the liver vessel centerline heatmap. An over-reconstructed liver vessel graph model is then traced based on the heatmap using an image processing based algorithm. We use a graph attention network to prune the false-positive branches by predicting the presence probability of each segmented branch in the initial reconstruction using the aggregated CNN features. We evaluated the proposed framework on an in-house dataset consisting of 418 multi-phase abdomen CT images with contrast. The proposed graph network pruning improves the overall reconstruction F1 score by 6.4% over the baseline. It also outperformed the other state-of-the-art curvilinear structure reconstruction algorithms.
Montage based 3D Medical Image Retrieval from Traumatic Brain Injury Cohort using Deep Convolutional Neural Network
Dec 10, 2018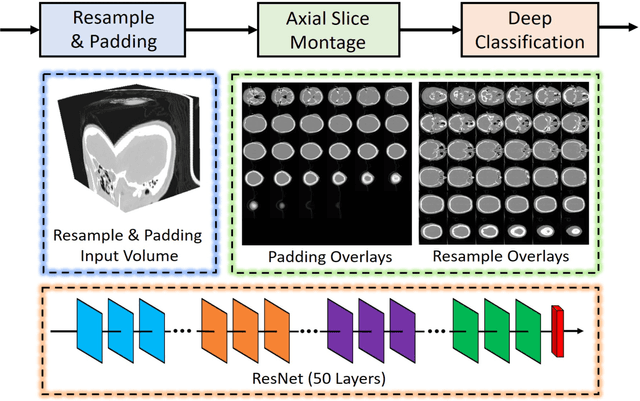
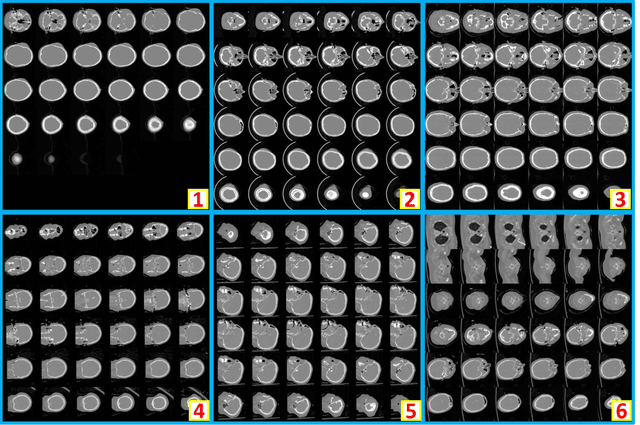
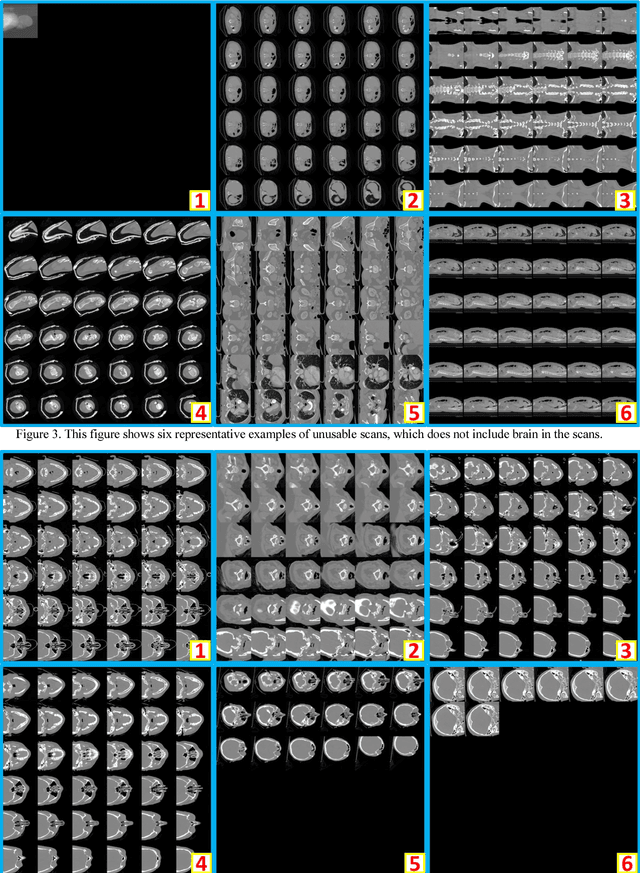
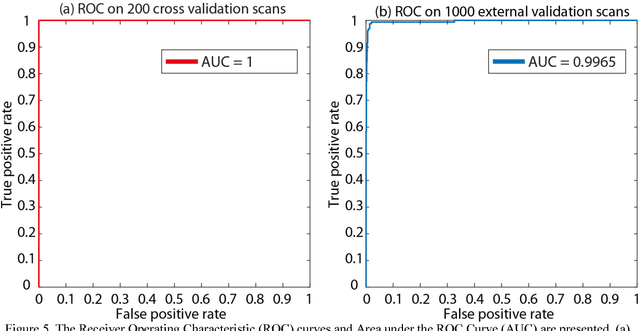
Abstract:Brain imaging analysis on clinically acquired computed tomography (CT) is essential for the diagnosis, risk prediction of progression, and treatment of the structural phenotypes of traumatic brain injury (TBI). However, in real clinical imaging scenarios, entire body CT images (e.g., neck, abdomen, chest, pelvis) are typically captured along with whole brain CT scans. For instance, in a typical sample of clinical TBI imaging cohort, only ~15% of CT scans actually contain whole brain CT images suitable for volumetric brain analyses; the remaining are partial brain or non-brain images. Therefore, a manual image retrieval process is typically required to isolate the whole brain CT scans from the entire cohort. However, the manual image retrieval is time and resource consuming and even more difficult for the larger cohorts. To alleviate the manual efforts, in this paper we propose an automated 3D medical image retrieval pipeline, called deep montage-based image retrieval (dMIR), which performs classification on 2D montage images via a deep convolutional neural network. The novelty of the proposed method for image processing is to characterize the medical image retrieval task based on the montage images. In a cohort of 2000 clinically acquired TBI scans, 794 scans were used as training data, 206 scans were used as validation data, and the remaining 1000 scans were used as testing data. The proposed achieved accuracy=1.0, recall=1.0, precision=1.0, f1=1.0 for validation data, while achieved accuracy=0.988, recall=0.962, precision=0.962, f1=0.962 for testing data. Thus, the proposed dMIR is able to perform accurate CT whole brain image retrieval from large-scale clinical cohorts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge