Donghao Zhang
EventRPG: Event Data Augmentation with Relevance Propagation Guidance
Mar 14, 2024



Abstract:Event camera, a novel bio-inspired vision sensor, has drawn a lot of attention for its low latency, low power consumption, and high dynamic range. Currently, overfitting remains a critical problem in event-based classification tasks for Spiking Neural Network (SNN) due to its relatively weak spatial representation capability. Data augmentation is a simple but efficient method to alleviate overfitting and improve the generalization ability of neural networks, and saliency-based augmentation methods are proven to be effective in the image processing field. However, there is no approach available for extracting saliency maps from SNNs. Therefore, for the first time, we present Spiking Layer-Time-wise Relevance Propagation rule (SLTRP) and Spiking Layer-wise Relevance Propagation rule (SLRP) in order for SNN to generate stable and accurate CAMs and saliency maps. Based on this, we propose EventRPG, which leverages relevance propagation on the spiking neural network for more efficient augmentation. Our proposed method has been evaluated on several SNN structures, achieving state-of-the-art performance in object recognition tasks including N-Caltech101, CIFAR10-DVS, with accuracies of 85.62% and 85.55%, as well as action recognition task SL-Animals with an accuracy of 91.59%. Our code is available at https://github.com/myuansun/EventRPG.
3D Matting: A Benchmark Study on Soft Segmentation Method for Pulmonary Nodules Applied in Computed Tomography
Oct 11, 2022



Abstract:Usually, lesions are not isolated but are associated with the surrounding tissues. For example, the growth of a tumour can depend on or infiltrate into the surrounding tissues. Due to the pathological nature of the lesions, it is challenging to distinguish their boundaries in medical imaging. However, these uncertain regions may contain diagnostic information. Therefore, the simple binarization of lesions by traditional binary segmentation can result in the loss of diagnostic information. In this work, we introduce the image matting into the 3D scenes and use the alpha matte, i.e., a soft mask, to describe lesions in a 3D medical image. The traditional soft mask acted as a training trick to compensate for the easily mislabelled or under-labelled ambiguous regions. In contrast, 3D matting uses soft segmentation to characterize the uncertain regions more finely, which means that it retains more structural information for subsequent diagnosis and treatment. The current study of image matting methods in 3D is limited. To address this issue, we conduct a comprehensive study of 3D matting, including both traditional and deep-learning-based methods. We adapt four state-of-the-art 2D image matting algorithms to 3D scenes and further customize the methods for CT images to calibrate the alpha matte with the radiodensity. Moreover, we propose the first end-to-end deep 3D matting network and implement a solid 3D medical image matting benchmark. Its efficient counterparts are also proposed to achieve a good performance-computation balance. Furthermore, there is no high-quality annotated dataset related to 3D matting, slowing down the development of data-driven deep-learning-based methods. To address this issue, we construct the first 3D medical matting dataset. The validity of the dataset was verified through clinicians' assessments and downstream experiments.
3D Matting: A Soft Segmentation Method Applied in Computed Tomography
Sep 16, 2022



Abstract:Three-dimensional (3D) images, such as CT, MRI, and PET, are common in medical imaging applications and important in clinical diagnosis. Semantic ambiguity is a typical feature of many medical image labels. It can be caused by many factors, such as the imaging properties, pathological anatomy, and the weak representation of the binary masks, which brings challenges to accurate 3D segmentation. In 2D medical images, using soft masks instead of binary masks generated by image matting to characterize lesions can provide rich semantic information, describe the structural characteristics of lesions more comprehensively, and thus benefit the subsequent diagnoses and analyses. In this work, we introduce image matting into the 3D scenes to describe the lesions in 3D medical images. The study of image matting in 3D modality is limited, and there is no high-quality annotated dataset related to 3D matting, therefore slowing down the development of data-driven deep-learning-based methods. To address this issue, we constructed the first 3D medical matting dataset and convincingly verified the validity of the dataset through quality control and downstream experiments in lung nodules classification. We then adapt the four selected state-of-the-art 2D image matting algorithms to 3D scenes and further customize the methods for CT images. Also, we propose the first end-to-end deep 3D matting network and implement a solid 3D medical image matting benchmark, which will be released to encourage further research.
Camera Adaptation for Fundus-Image-Based CVD Risk Estimation
Jun 18, 2022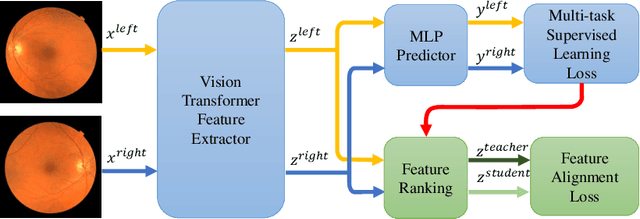
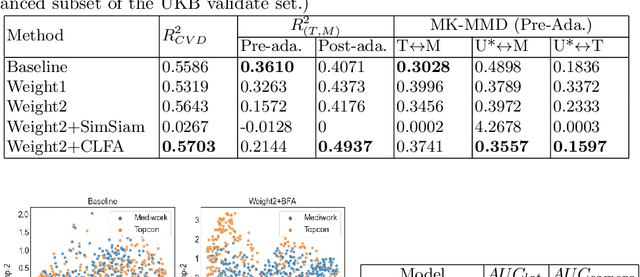
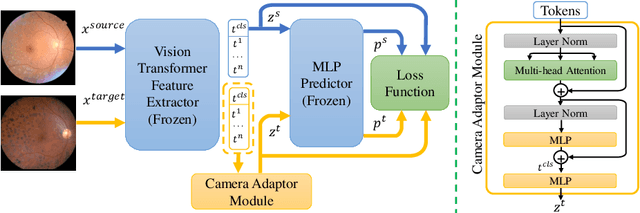

Abstract:Recent studies have validated the association between cardiovascular disease (CVD) risk and retinal fundus images. Combining deep learning (DL) and portable fundus cameras will enable CVD risk estimation in various scenarios and improve healthcare democratization. However, there are still significant issues to be solved. One of the top priority issues is the different camera differences between the databases for research material and the samples in the production environment. Most high-quality retinography databases ready for research are collected from high-end fundus cameras, and there is a significant domain discrepancy between different cameras. To fully explore the domain discrepancy issue, we first collect a Fundus Camera Paired (FCP) dataset containing pair-wise fundus images captured by the high-end Topcon retinal camera and the low-end Mediwork portable fundus camera of the same patients. Then, we propose a cross-laterality feature alignment pre-training scheme and a self-attention camera adaptor module to improve the model robustness. The cross-laterality feature alignment training encourages the model to learn common knowledge from the same patient's left and right fundus images and improve model generalization. Meanwhile, the device adaptation module learns feature transformation from the target domain to the source domain. We conduct comprehensive experiments on both the UK Biobank database and our FCP data. The experimental results show that the CVD risk regression accuracy and the result consistency over two cameras are improved with our proposed method. The code is available here: \url{https://github.com/linzhlalala/CVD-risk-based-on-retinal-fundus-images}
Medical Visual Question Answering: A Survey
Nov 19, 2021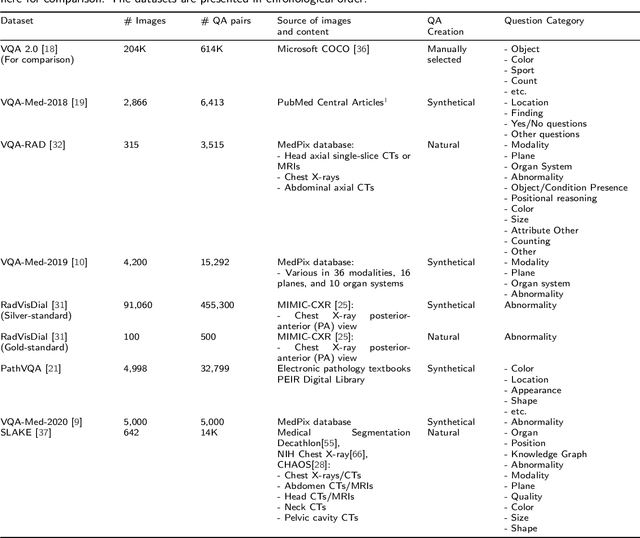
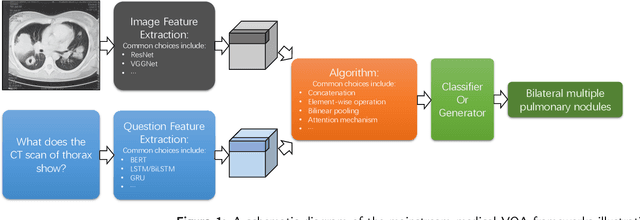
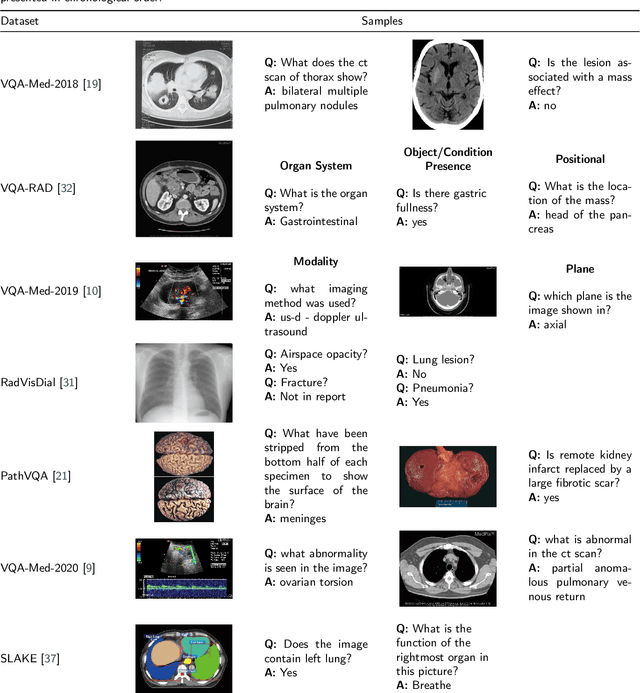
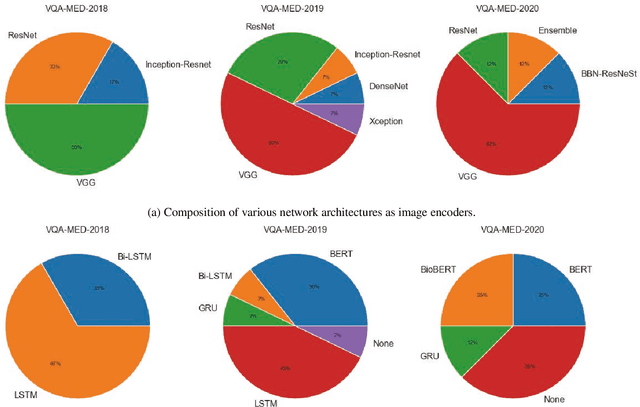
Abstract:Medical Visual Question Answering (VQA) is a combination of medical artificial intelligence and popular VQA challenges. Given a medical image and a clinically relevant question in natural language, the medical VQA system is expected to predict a plausible and convincing answer. Although the general-domain VQA has been extensively studied, the medical VQA still needs specific investigation and exploration due to its task features. In the first part of this survey, we cover and discuss the publicly available medical VQA datasets up to date about the data source, data quantity, and task feature. In the second part, we review the approaches used in medical VQA tasks. In the last part, we analyze some medical-specific challenges for the field and discuss future research directions.
Enforcing Mutual Consistency of Hard Regions for Semi-supervised Medical Image Segmentation
Sep 21, 2021



Abstract:In this paper, we proposed a novel mutual consistency network (MC-Net+) to effectively exploit the unlabeled hard regions for semi-supervised medical image segmentation. The MC-Net+ model is motivated by the observation that deep models trained with limited annotations are prone to output highly uncertain and easily mis-classified predictions in the ambiguous regions (e.g. adhesive edges or thin branches) for the image segmentation task. Leveraging these region-level challenging samples can make the semi-supervised segmentation model training more effective. Therefore, our proposed MC-Net+ model consists of two new designs. First, the model contains one shared encoder and multiple sightly different decoders (i.e. using different up-sampling strategies). The statistical discrepancy of multiple decoders' outputs is computed to denote the model's uncertainty, which indicates the unlabeled hard regions. Second, a new mutual consistency constraint is enforced between one decoder's probability output and other decoders' soft pseudo labels. In this way, we minimize the model's uncertainty during training and force the model to generate invariant and low-entropy results in such challenging areas of unlabeled data, in order to learn a generalized feature representation. We compared the segmentation results of the MC-Net+ with five state-of-the-art semi-supervised approaches on three public medical datasets. Extension experiments with two common semi-supervised settings demonstrate the superior performance of our model over other existing methods, which sets a new state of the art for semi-supervised medical image segmentation.
Medical Matting: A New Perspective on Medical Segmentation with Uncertainty
Jul 08, 2021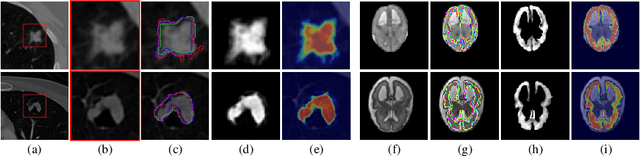
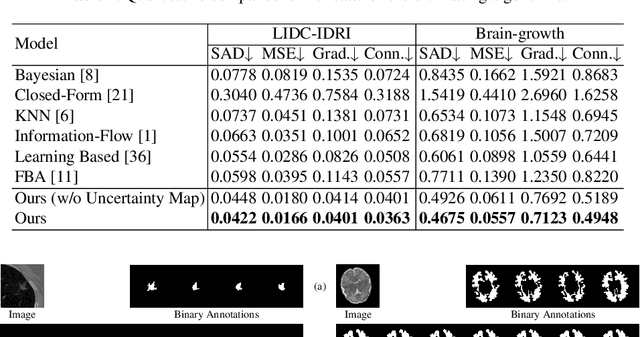

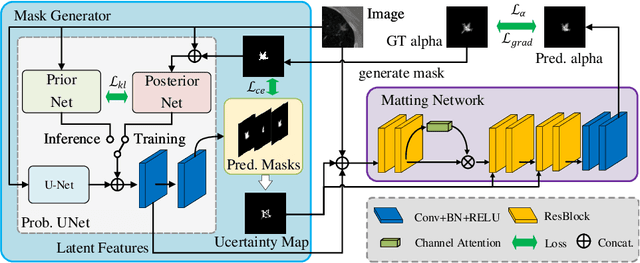
Abstract:In medical image segmentation, it is difficult to mark ambiguous areas accurately with binary masks, especially when dealing with small lesions. Therefore, it is a challenge for radiologists to reach a consensus by using binary masks under the condition of multiple annotations. However, these areas may contain anatomical structures that are conducive to diagnosis. Uncertainty is introduced to study these situations. Nevertheless, the uncertainty is usually measured by the variances between predictions in a multiple trial way. It is not intuitive, and there is no exact correspondence in the image. Inspired by image matting, we introduce matting as a soft segmentation method and a new perspective to deal with and represent uncertain regions into medical scenes, namely medical matting. More specifically, because there is no available medical matting dataset, we first labeled two medical datasets with alpha matte. Secondly, the matting method applied to the natural image is not suitable for the medical scene, so we propose a new architecture to generate binary masks and alpha matte in a row. Thirdly, the uncertainty map is introduced to highlight the ambiguous regions from the binary results and improve the matting performance. Evaluated on these datasets, the proposed model outperformed state-of-the-art matting algorithms by a large margin, and alpha matte is proved to be a more efficient labeling form than a binary mask.
Unsupervised Instance Segmentation in Microscopy Images via Panoptic Domain Adaptation and Task Re-weighting
May 05, 2020



Abstract:Unsupervised domain adaptation (UDA) for nuclei instance segmentation is important for digital pathology, as it alleviates the burden of labor-intensive annotation and domain shift across datasets. In this work, we propose a Cycle Consistency Panoptic Domain Adaptive Mask R-CNN (CyC-PDAM) architecture for unsupervised nuclei segmentation in histopathology images, by learning from fluorescence microscopy images. More specifically, we first propose a nuclei inpainting mechanism to remove the auxiliary generated objects in the synthesized images. Secondly, a semantic branch with a domain discriminator is designed to achieve panoptic-level domain adaptation. Thirdly, in order to avoid the influence of the source-biased features, we propose a task re-weighting mechanism to dynamically add trade-off weights for the task-specific loss functions. Experimental results on three datasets indicate that our proposed method outperforms state-of-the-art UDA methods significantly, and demonstrates a similar performance as fully supervised methods.
Graph Attention Network based Pruning for Reconstructing 3D Liver Vessel Morphology from Contrasted CT Images
Mar 18, 2020



Abstract:With the injection of contrast material into blood vessels, multi-phase contrasted CT images can enhance the visibility of vessel networks in the human body. Reconstructing the 3D geometric morphology of liver vessels from the contrasted CT images can enable multiple liver preoperative surgical planning applications. Automatic reconstruction of liver vessel morphology remains a challenging problem due to the morphological complexity of liver vessels and the inconsistent vessel intensities among different multi-phase contrasted CT images. On the other side, high integrity is required for the 3D reconstruction to avoid decision making biases. In this paper, we propose a framework for liver vessel morphology reconstruction using both a fully convolutional neural network and a graph attention network. A fully convolutional neural network is first trained to produce the liver vessel centerline heatmap. An over-reconstructed liver vessel graph model is then traced based on the heatmap using an image processing based algorithm. We use a graph attention network to prune the false-positive branches by predicting the presence probability of each segmented branch in the initial reconstruction using the aggregated CNN features. We evaluated the proposed framework on an in-house dataset consisting of 418 multi-phase abdomen CT images with contrast. The proposed graph network pruning improves the overall reconstruction F1 score by 6.4% over the baseline. It also outperformed the other state-of-the-art curvilinear structure reconstruction algorithms.
Cell R-CNN V3: A Novel Panoptic Paradigm for Instance Segmentation in Biomedical Images
Feb 15, 2020



Abstract:Instance segmentation is an important task for biomedical image analysis. Due to the complicated background components, the high variability of object appearances, numerous overlapping objects, and ambiguous object boundaries, this task still remains challenging. Recently, deep learning based methods have been widely employed to solve these problems and can be categorized into proposal-free and proposal-based methods. However, both proposal-free and proposal-based methods suffer from information loss, as they focus on either global-level semantic or local-level instance features. To tackle this issue, we present a panoptic architecture that unifies the semantic and instance features in this work. Specifically, our proposed method contains a residual attention feature fusion mechanism to incorporate the instance prediction with the semantic features, in order to facilitate the semantic contextual information learning in the instance branch. Then, a mask quality branch is designed to align the confidence score of each object with the quality of the mask prediction. Furthermore, a consistency regularization mechanism is designed between the semantic segmentation tasks in the semantic and instance branches, for the robust learning of both tasks. Extensive experiments demonstrate the effectiveness of our proposed method, which outperforms several state-of-the-art methods on various biomedical datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge