Birgi Tamersoy
Automated CT Lung Cancer Screening Workflow using 3D Camera
Sep 27, 2023Abstract:Despite recent developments in CT planning that enabled automation in patient positioning, time-consuming scout scans are still needed to compute dose profile and ensure the patient is properly positioned. In this paper, we present a novel method which eliminates the need for scout scans in CT lung cancer screening by estimating patient scan range, isocenter, and Water Equivalent Diameter (WED) from 3D camera images. We achieve this task by training an implicit generative model on over 60,000 CT scans and introduce a novel approach for updating the prediction using real-time scan data. We demonstrate the effectiveness of our method on a testing set of 110 pairs of depth data and CT scan, resulting in an average error of 5mm in estimating the isocenter, 13mm in determining the scan range, 10mm and 16mm in estimating the AP and lateral WED respectively. The relative WED error of our method is 4%, which is well within the International Electrotechnical Commission (IEC) acceptance criteria of 10%.
Adaloss: Adaptive Loss Function for Landmark Localization
Aug 02, 2019



Abstract:Landmark localization is a challenging problem in computer vision with a multitude of applications. Recent deep learning based methods have shown improved results by regressing likelihood maps instead of regressing the coordinates directly. However, setting the precision of these regression targets during the training is a cumbersome process since it creates a trade-off between trainability vs localization accuracy. Using precise targets introduces a significant sampling bias and hence makes the training more difficult, whereas using imprecise targets results in inaccurate landmark detectors. In this paper, we introduce "Adaloss", an objective function that adapts itself during the training by updating the target precision based on the training statistics. This approach does not require setting problem-specific parameters and shows improved stability in training and better localization accuracy during inference. We demonstrate the effectiveness of our proposed method in three different applications of landmark localization: 1) the challenging task of precisely detecting catheter tips in medical X-ray images, 2) localizing surgical instruments in endoscopic images, and 3) localizing facial features on in-the-wild images where we show state-of-the-art results on the 300-W benchmark dataset.
Generating Synthetic X-ray Images of a Person from the Surface Geometry
May 14, 2018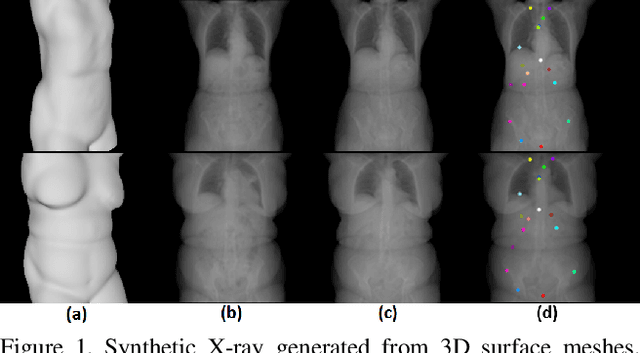
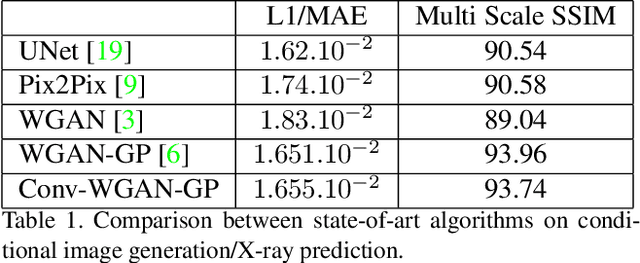
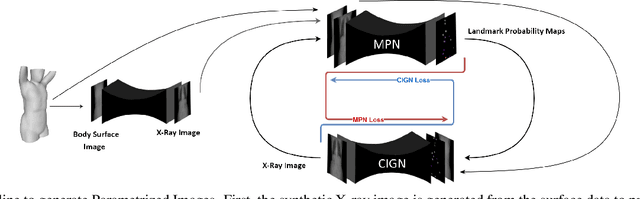
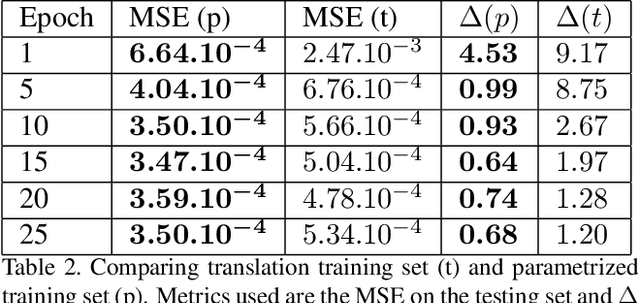
Abstract:We present a novel framework that learns to predict human anatomy from body surface. Specifically, our approach generates a synthetic X-ray image of a person only from the person's surface geometry. Furthermore, the synthetic X-ray image is parametrized and can be manipulated by adjusting a set of body markers which are also generated during the X-ray image prediction. With the proposed framework, multiple synthetic X-ray images can easily be generated by varying surface geometry. By perturbing the parameters, several additional synthetic X-ray images can be generated from the same surface geometry. As a result, our approach offers a potential to overcome the training data barrier in the medical domain. This capability is achieved by learning a pair of networks - one learns to generate the full image from the partial image and a set of parameters, and the other learns to estimate the parameters given the full image. During training, the two networks are trained iteratively such that they would converge to a solution where the predicted parameters and the full image are consistent with each other. In addition to medical data enrichment, our framework can also be used for image completion as well as anomaly detection.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge