Ugur Demir
Emulators for stellar profiles in binary population modeling
Oct 14, 2024



Abstract:Knowledge about the internal physical structure of stars is crucial to understanding their evolution. The novel binary population synthesis code POSYDON includes a module for interpolating the stellar and binary properties of any system at the end of binary MESA evolution based on a pre-computed set of models. In this work, we present a new emulation method for predicting stellar profiles, i.e., the internal stellar structure along the radial axis, using machine learning techniques. We use principal component analysis for dimensionality reduction and fully-connected feed-forward neural networks for making predictions. We find accuracy to be comparable to that of nearest neighbor approximation, with a strong advantage in terms of memory and storage efficiency. By delivering more information about the evolution of stellar internal structure, these emulators will enable faster simulations of higher physical fidelity with large-scale simulations of binary star population synthesis possible with POSYDON and other population synthesis codes.
Explainable Transformer Prototypes for Medical Diagnoses
Mar 11, 2024


Abstract:Deployments of artificial intelligence in medical diagnostics mandate not just accuracy and efficacy but also trust, emphasizing the need for explainability in machine decisions. The recent trend in automated medical image diagnostics leans towards the deployment of Transformer-based architectures, credited to their impressive capabilities. Since the self-attention feature of transformers contributes towards identifying crucial regions during the classification process, they enhance the trustability of the methods. However, the complex intricacies of these attention mechanisms may fall short of effectively pinpointing the regions of interest directly influencing AI decisions. Our research endeavors to innovate a unique attention block that underscores the correlation between 'regions' rather than 'pixels'. To address this challenge, we introduce an innovative system grounded in prototype learning, featuring an advanced self-attention mechanism that goes beyond conventional ad-hoc visual explanation techniques by offering comprehensible visual insights. A combined quantitative and qualitative methodological approach was used to demonstrate the effectiveness of the proposed method on the large-scale NIH chest X-ray dataset. Experimental results showed that our proposed method offers a promising direction for explainability, which can lead to the development of more trustable systems, which can facilitate easier and rapid adoption of such technology into routine clinics. The code is available at www.github.com/NUBagcilab/r2r_proto.
Radiomics Boosts Deep Learning Model for IPMN Classification
Sep 11, 2023Abstract:Intraductal Papillary Mucinous Neoplasm (IPMN) cysts are pre-malignant pancreas lesions, and they can progress into pancreatic cancer. Therefore, detecting and stratifying their risk level is of ultimate importance for effective treatment planning and disease control. However, this is a highly challenging task because of the diverse and irregular shape, texture, and size of the IPMN cysts as well as the pancreas. In this study, we propose a novel computer-aided diagnosis pipeline for IPMN risk classification from multi-contrast MRI scans. Our proposed analysis framework includes an efficient volumetric self-adapting segmentation strategy for pancreas delineation, followed by a newly designed deep learning-based classification scheme with a radiomics-based predictive approach. We test our proposed decision-fusion model in multi-center data sets of 246 multi-contrast MRI scans and obtain superior performance to the state of the art (SOTA) in this field. Our ablation studies demonstrate the significance of both radiomics and deep learning modules for achieving the new SOTA performance compared to international guidelines and published studies (81.9\% vs 61.3\% in accuracy). Our findings have important implications for clinical decision-making. In a series of rigorous experiments on multi-center data sets (246 MRI scans from five centers), we achieved unprecedented performance (81.9\% accuracy).
Ensuring Trustworthy Medical Artificial Intelligence through Ethical and Philosophical Principles
Apr 29, 2023Abstract:Artificial intelligence (AI) methods have great potential to revolutionize numerous medical care by enhancing the experience of medical experts and patients. AI based computer-assisted diagnosis tools can have a tremendous benefit if they can outperform or perform similarly to the level of a clinical expert. As a result, advanced healthcare services can be affordable in developing nations, and the problem of a lack of expert medical practitioners can be addressed. AI based tools can save time, resources, and overall cost for patient treatment. Furthermore, in contrast to humans, AI can uncover complex relations in the data from a large set of inputs and even lead to new evidence-based knowledge in medicine. However, integrating AI in healthcare raises several ethical and philosophical concerns, such as bias, transparency, autonomy, responsibility and accountability, which must be addressed before integrating such tools into clinical settings. In this article, we emphasize recent advances in AI-assisted medical image analysis, existing standards, and the significance of comprehending ethical issues and best practices for the applications of AI in clinical settings. We cover the technical and ethical challenges of AI and the implications of deploying AI in hospitals and public organizations. We also discuss promising key measures and techniques to address the ethical challenges, data scarcity, racial bias, lack of transparency, and algorithmic bias. Finally, we provide our recommendation and future directions for addressing the ethical challenges associated with AI in healthcare applications, with the goal of deploying AI into the clinical settings to make the workflow more efficient, accurate, accessible, transparent, and reliable for the patient worldwide.
Domain Generalization with Adversarial Intensity Attack for Medical Image Segmentation
Apr 05, 2023Abstract:Most statistical learning algorithms rely on an over-simplified assumption, that is, the train and test data are independent and identically distributed. In real-world scenarios, however, it is common for models to encounter data from new and different domains to which they were not exposed to during training. This is often the case in medical imaging applications due to differences in acquisition devices, imaging protocols, and patient characteristics. To address this problem, domain generalization (DG) is a promising direction as it enables models to handle data from previously unseen domains by learning domain-invariant features robust to variations across different domains. To this end, we introduce a novel DG method called Adversarial Intensity Attack (AdverIN), which leverages adversarial training to generate training data with an infinite number of styles and increase data diversity while preserving essential content information. We conduct extensive evaluation experiments on various multi-domain segmentation datasets, including 2D retinal fundus optic disc/cup and 3D prostate MRI. Our results demonstrate that AdverIN significantly improves the generalization ability of the segmentation models, achieving significant improvement on these challenging datasets. Code is available upon publication.
Domain Generalization with Correlated Style Uncertainty
Dec 20, 2022Abstract:Though impressive success has been witnessed in computer vision, deep learning still suffers from the domain shift challenge when the target domain for testing and the source domain for training do not share an identical distribution. To address this, domain generalization approaches intend to extract domain invariant features that can lead to a more robust model. Hence, increasing the source domain diversity is a key component of domain generalization. Style augmentation takes advantage of instance-specific feature statistics containing informative style characteristics to synthetic novel domains. However, all previous works ignored the correlation between different feature channels or only limited the style augmentation through linear interpolation. In this work, we propose a novel augmentation method, called \textit{Correlated Style Uncertainty (CSU)}, to go beyond the linear interpolation of style statistic space while preserving the essential correlation information. We validate our method's effectiveness by extensive experiments on multiple cross-domain classification tasks, including widely used PACS, Office-Home, Camelyon17 datasets and the Duke-Market1501 instance retrieval task and obtained significant margin improvements over the state-of-the-art methods. The source code is available for public use.
A Critical Appraisal of Data Augmentation Methods for Imaging-Based Medical Diagnosis Applications
Dec 14, 2022

Abstract:Current data augmentation techniques and transformations are well suited for improving the size and quality of natural image datasets but are not yet optimized for medical imaging. We hypothesize that sub-optimal data augmentations can easily distort or occlude medical images, leading to false positives or negatives during patient diagnosis, prediction, or therapy/surgery evaluation. In our experimental results, we found that utilizing commonly used intensity-based data augmentation distorts the MRI scans and leads to texture information loss, thus negatively affecting the overall performance of classification. Additionally, we observed that commonly used data augmentation methods cannot be used with a plug-and-play approach in medical imaging, and requires manual tuning and adjustment.
Transformer based Generative Adversarial Network for Liver Segmentation
May 28, 2022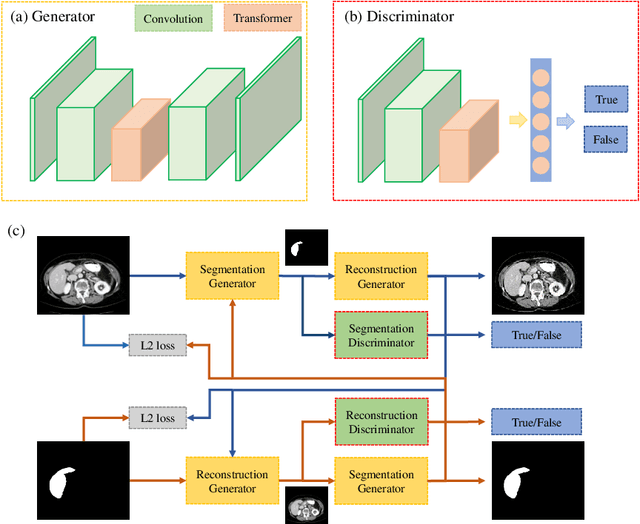
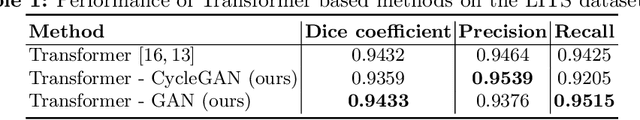
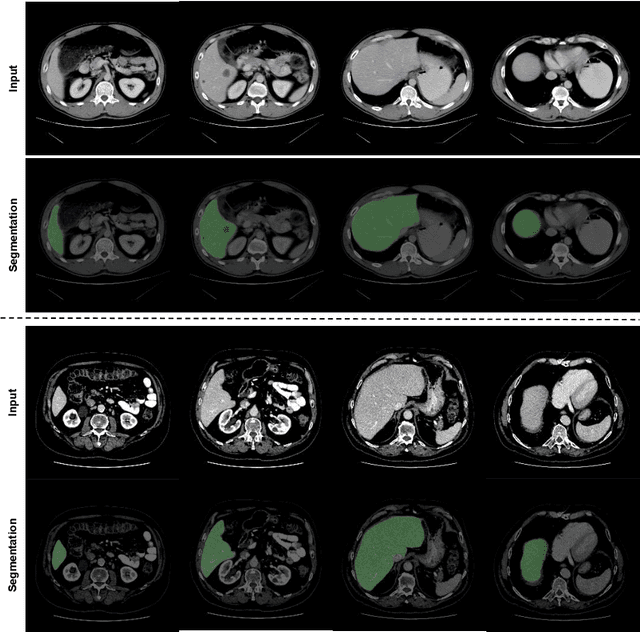
Abstract:Automated liver segmentation from radiology scans (CT, MRI) can improve surgery and therapy planning and follow-up assessment in addition to conventional use for diagnosis and prognosis. Although convolutional neural networks (CNNs) have become the standard image segmentation tasks, more recently this has started to change towards Transformers based architectures because Transformers are taking advantage of capturing long range dependence modeling capability in signals, so called attention mechanism. In this study, we propose a new segmentation approach using a hybrid approach combining the Transformer(s) with the Generative Adversarial Network (GAN) approach. The premise behind this choice is that the self-attention mechanism of the Transformers allows the network to aggregate the high dimensional feature and provide global information modeling. This mechanism provides better segmentation performance compared with traditional methods. Furthermore, we encode this generator into the GAN based architecture so that the discriminator network in the GAN can classify the credibility of the generated segmentation masks compared with the real masks coming from human (expert) annotations. This allows us to extract the high dimensional topology information in the mask for biomedical image segmentation and provide more reliable segmentation results. Our model achieved a high dice coefficient of 0.9433, recall of 0.9515, and precision of 0.9376 and outperformed other Transformer based approaches.
MFR 2021: Masked Face Recognition Competition
Jun 29, 2021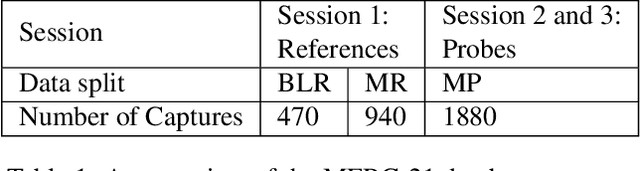

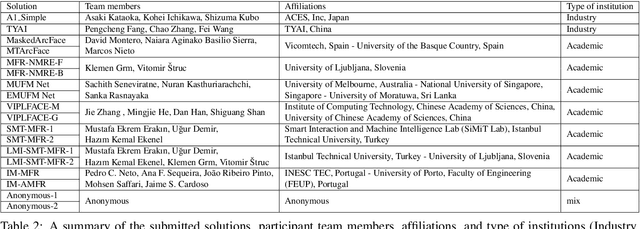
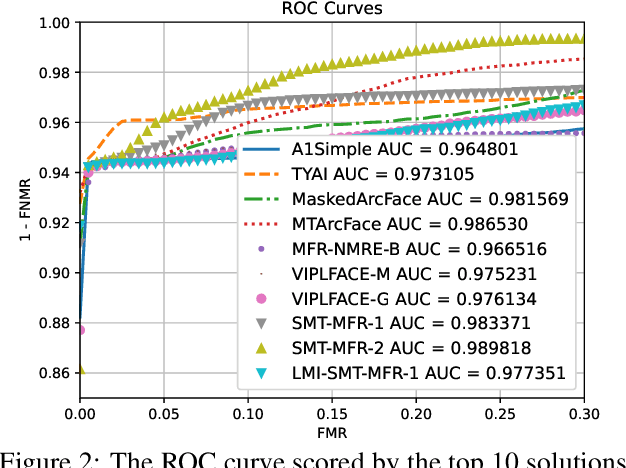
Abstract:This paper presents a summary of the Masked Face Recognition Competitions (MFR) held within the 2021 International Joint Conference on Biometrics (IJCB 2021). The competition attracted a total of 10 participating teams with valid submissions. The affiliations of these teams are diverse and associated with academia and industry in nine different countries. These teams successfully submitted 18 valid solutions. The competition is designed to motivate solutions aiming at enhancing the face recognition accuracy of masked faces. Moreover, the competition considered the deployability of the proposed solutions by taking the compactness of the face recognition models into account. A private dataset representing a collaborative, multi-session, real masked, capture scenario is used to evaluate the submitted solutions. In comparison to one of the top-performing academic face recognition solutions, 10 out of the 18 submitted solutions did score higher masked face verification accuracy.
Information Bottleneck Attribution for Visual Explanations of Diagnosis and Prognosis
Apr 07, 2021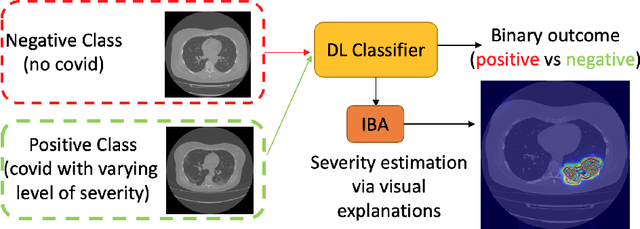



Abstract:Visual explanation methods have an important role in the prognosis of the patients where the annotated data is limited or not available. There have been several attempts to use gradient-based attribution methods to localize pathology from medical scans without using segmentation labels. This research direction has been impeded by the lack of robustness and reliability. These methods are highly sensitive to the network parameters. In this study, we introduce a robust visual explanation method to address this problem for medical applications. We provide a highly innovative algorithm to quantifying lesions in the lungs caused by the Covid-19 with high accuracy and robustness without using dense segmentation labels. Inspired by the information bottleneck concept, we mask the neural network representation with noise to find out important regions. This approach overcomes the drawbacks of commonly used Grad-Cam and its derived algorithms. The premise behind our proposed strategy is that the information flow is minimized while ensuring the classifier prediction stays similar. Our findings indicate that the bottleneck condition provides a more stable and robust severity estimation than the similar attribution methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge