Michael Wallace
Challenges for Predictive Modeling with Neural Network Techniques using Error-Prone Dietary Intake Data
Nov 15, 2023



Abstract:Dietary intake data are routinely drawn upon to explore diet-health relationships. However, these data are often subject to measurement error, distorting the true relationships. Beyond measurement error, there are likely complex synergistic and sometimes antagonistic interactions between different dietary components, complicating the relationships between diet and health outcomes. Flexible models are required to capture the nuance that these complex interactions introduce. This complexity makes research on diet-health relationships an appealing candidate for the application of machine learning techniques, and in particular, neural networks. Neural networks are computational models that are able to capture highly complex, nonlinear relationships so long as sufficient data are available. While these models have been applied in many domains, the impacts of measurement error on the performance of predictive modeling has not been systematically investigated. However, dietary intake data are typically collected using self-report methods and are prone to large amounts of measurement error. In this work, we demonstrate the ways in which measurement error erodes the performance of neural networks, and illustrate the care that is required for leveraging these models in the presence of error. We demonstrate the role that sample size and replicate measurements play on model performance, indicate a motivation for the investigation of transformations to additivity, and illustrate the caution required to prevent model overfitting. While the past performance of neural networks across various domains make them an attractive candidate for examining diet-health relationships, our work demonstrates that substantial care and further methodological development are both required to observe increased predictive performance when applying these techniques, compared to more traditional statistical procedures.
Radiomics Boosts Deep Learning Model for IPMN Classification
Sep 11, 2023Abstract:Intraductal Papillary Mucinous Neoplasm (IPMN) cysts are pre-malignant pancreas lesions, and they can progress into pancreatic cancer. Therefore, detecting and stratifying their risk level is of ultimate importance for effective treatment planning and disease control. However, this is a highly challenging task because of the diverse and irregular shape, texture, and size of the IPMN cysts as well as the pancreas. In this study, we propose a novel computer-aided diagnosis pipeline for IPMN risk classification from multi-contrast MRI scans. Our proposed analysis framework includes an efficient volumetric self-adapting segmentation strategy for pancreas delineation, followed by a newly designed deep learning-based classification scheme with a radiomics-based predictive approach. We test our proposed decision-fusion model in multi-center data sets of 246 multi-contrast MRI scans and obtain superior performance to the state of the art (SOTA) in this field. Our ablation studies demonstrate the significance of both radiomics and deep learning modules for achieving the new SOTA performance compared to international guidelines and published studies (81.9\% vs 61.3\% in accuracy). Our findings have important implications for clinical decision-making. In a series of rigorous experiments on multi-center data sets (246 MRI scans from five centers), we achieved unprecedented performance (81.9\% accuracy).
A Critical Appraisal of Data Augmentation Methods for Imaging-Based Medical Diagnosis Applications
Dec 14, 2022

Abstract:Current data augmentation techniques and transformations are well suited for improving the size and quality of natural image datasets but are not yet optimized for medical imaging. We hypothesize that sub-optimal data augmentations can easily distort or occlude medical images, leading to false positives or negatives during patient diagnosis, prediction, or therapy/surgery evaluation. In our experimental results, we found that utilizing commonly used intensity-based data augmentation distorts the MRI scans and leads to texture information loss, thus negatively affecting the overall performance of classification. Additionally, we observed that commonly used data augmentation methods cannot be used with a plug-and-play approach in medical imaging, and requires manual tuning and adjustment.
PAN: Projective Adversarial Network for Medical Image Segmentation
Jun 11, 2019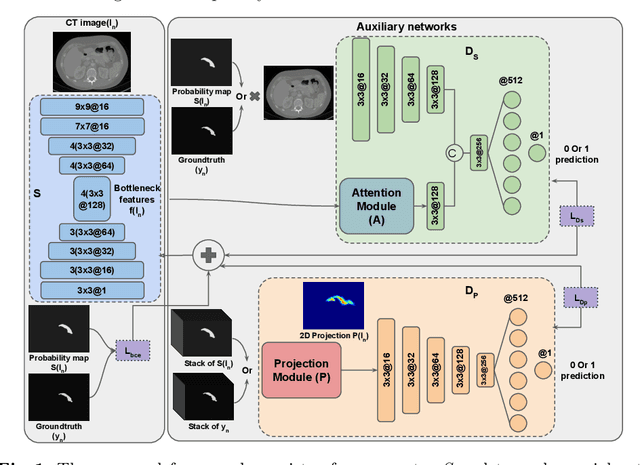
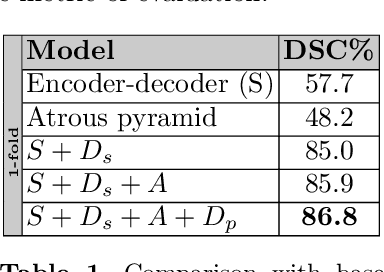
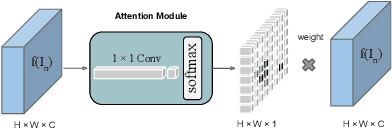
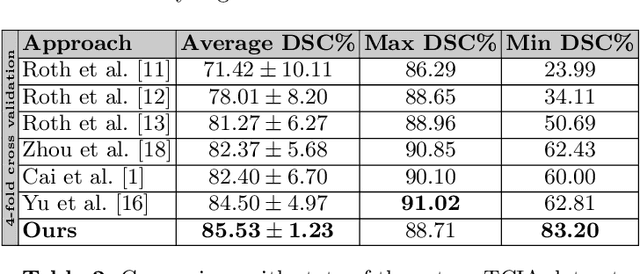
Abstract:Adversarial learning has been proven to be effective for capturing long-range and high-level label consistencies in semantic segmentation. Unique to medical imaging, capturing 3D semantics in an effective yet computationally efficient way remains an open problem. In this study, we address this computational burden by proposing a novel projective adversarial network, called PAN, which incorporates high-level 3D information through 2D projections. Furthermore, we introduce an attention module into our framework that helps for a selective integration of global information directly from our segmentor to our adversarial network. For the clinical application we chose pancreas segmentation from CT scans. Our proposed framework achieved state-of-the-art performance without adding to the complexity of the segmentor.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge