Paul Fieguth
Joint Embedding Variational Bayes
Feb 05, 2026Abstract:We introduce Variational Joint Embedding (VJE), a framework that synthesizes joint embedding and variational inference to enable self-supervised learning of probabilistic representations in a reconstruction-free, non-contrastive setting. Compared to energy-based predictive objectives that optimize pointwise discrepancies, VJE maximizes a symmetric conditional evidence lower bound (ELBO) for a latent-variable model defined directly on encoder embeddings. We instantiate the conditional likelihood with a heavy-tailed Student-$t$ model using a polar decomposition that explicitly decouples directional and radial factors to prevent norm-induced instabilities during training. VJE employs an amortized inference network to parameterize a diagonal Gaussian variational posterior whose feature-wise variances are shared with the likelihood scale to capture anisotropic uncertainty without auxiliary projection heads. Across ImageNet-1K, CIFAR-10/100, and STL-10, VJE achieves performance comparable to standard non-contrastive baselines under linear and k-NN evaluation. We further validate these probabilistic semantics through one-class CIFAR-10 anomaly detection, where likelihood-based scoring under the proposed model outperforms comparable self-supervised baselines.
Kernel VICReg for Self-Supervised Learning in Reproducing Kernel Hilbert Space
Sep 08, 2025Abstract:Self-supervised learning (SSL) has emerged as a powerful paradigm for representation learning by optimizing geometric objectives--such as invariance to augmentations, variance preservation, and feature decorrelation--without requiring labels. However, most existing methods operate in Euclidean space, limiting their ability to capture nonlinear dependencies and geometric structures. In this work, we propose Kernel VICReg, a novel self-supervised learning framework that lifts the VICReg objective into a Reproducing Kernel Hilbert Space (RKHS). By kernelizing each term of the loss-variance, invariance, and covariance--we obtain a general formulation that operates on double-centered kernel matrices and Hilbert-Schmidt norms, enabling nonlinear feature learning without explicit mappings. We demonstrate that Kernel VICReg not only avoids representational collapse but also improves performance on tasks with complex or small-scale data. Empirical evaluations across MNIST, CIFAR-10, STL-10, TinyImageNet, and ImageNet100 show consistent gains over Euclidean VICReg, with particularly strong improvements on datasets where nonlinear structures are prominent. UMAP visualizations further confirm that kernel-based embeddings exhibit better isometry and class separation. Our results suggest that kernelizing SSL objectives is a promising direction for bridging classical kernel methods with modern representation learning.
Self-Supervised Learning Using Nonlinear Dependence
Jan 31, 2025



Abstract:Self-supervised learning has gained significant attention in contemporary applications, particularly due to the scarcity of labeled data. While existing SSL methodologies primarily address feature variance and linear correlations, they often neglect the intricate relations between samples and the nonlinear dependencies inherent in complex data. In this paper, we introduce Correlation-Dependence Self-Supervised Learning (CDSSL), a novel framework that unifies and extends existing SSL paradigms by integrating both linear correlations and nonlinear dependencies, encapsulating sample-wise and feature-wise interactions. Our approach incorporates the Hilbert-Schmidt Independence Criterion (HSIC) to robustly capture nonlinear dependencies within a Reproducing Kernel Hilbert Space, enriching representation learning. Experimental evaluations on diverse benchmarks demonstrate the efficacy of CDSSL in improving representation quality.
Particle-Filtering-based Latent Diffusion for Inverse Problems
Aug 25, 2024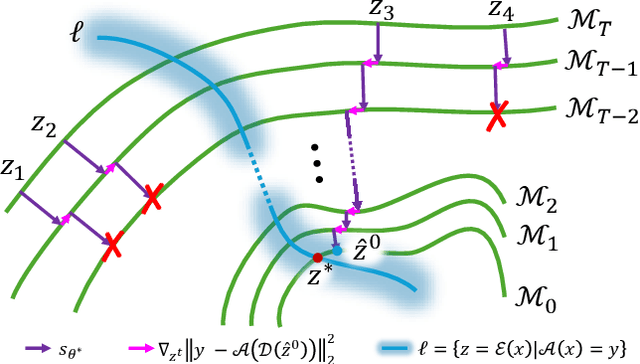
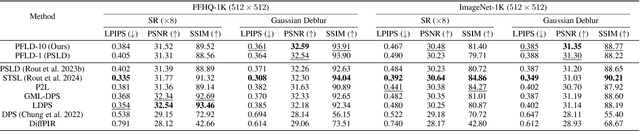
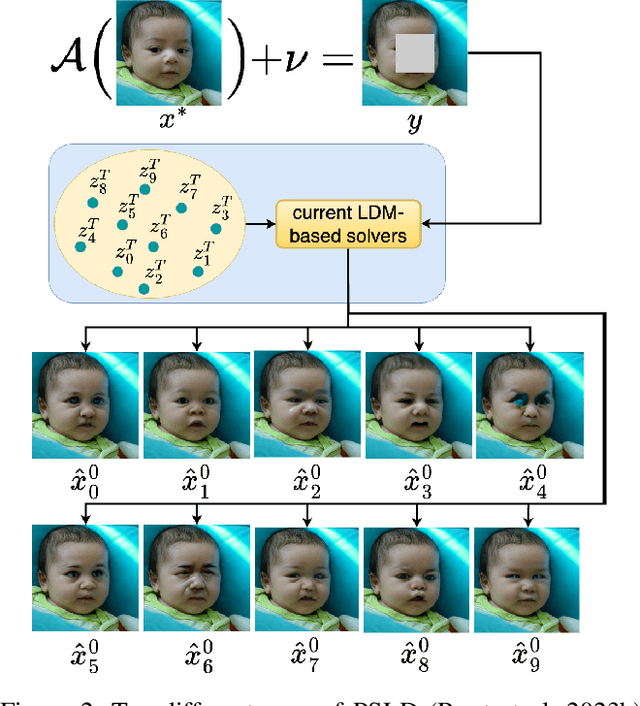

Abstract:Current strategies for solving image-based inverse problems apply latent diffusion models to perform posterior sampling.However, almost all approaches make no explicit attempt to explore the solution space, instead drawing only a single sample from a Gaussian distribution from which to generate their solution. In this paper, we introduce a particle-filtering-based framework for a nonlinear exploration of the solution space in the initial stages of reverse SDE methods. Our proposed particle-filtering-based latent diffusion (PFLD) method and proposed problem formulation and framework can be applied to any diffusion-based solution for linear or nonlinear inverse problems. Our experimental results show that PFLD outperforms the SoTA solver PSLD on the FFHQ-1K and ImageNet-1K datasets on inverse problem tasks of super resolution, Gaussian debluring and inpainting.
BIOSCAN-5M: A Multimodal Dataset for Insect Biodiversity
Jun 18, 2024



Abstract:As part of an ongoing worldwide effort to comprehend and monitor insect biodiversity, this paper presents the BIOSCAN-5M Insect dataset to the machine learning community and establish several benchmark tasks. BIOSCAN-5M is a comprehensive dataset containing multi-modal information for over 5 million insect specimens, and it significantly expands existing image-based biological datasets by including taxonomic labels, raw nucleotide barcode sequences, assigned barcode index numbers, and geographical information. We propose three benchmark experiments to demonstrate the impact of the multi-modal data types on the classification and clustering accuracy. First, we pretrain a masked language model on the DNA barcode sequences of the \mbox{BIOSCAN-5M} dataset, and demonstrate the impact of using this large reference library on species- and genus-level classification performance. Second, we propose a zero-shot transfer learning task applied to images and DNA barcodes to cluster feature embeddings obtained from self-supervised learning, to investigate whether meaningful clusters can be derived from these representation embeddings. Third, we benchmark multi-modality by performing contrastive learning on DNA barcodes, image data, and taxonomic information. This yields a general shared embedding space enabling taxonomic classification using multiple types of information and modalities. The code repository of the BIOSCAN-5M Insect dataset is available at {\url{https://github.com/zahrag/BIOSCAN-5M}}
Video Relationship Detection Using Mixture of Experts
Mar 06, 2024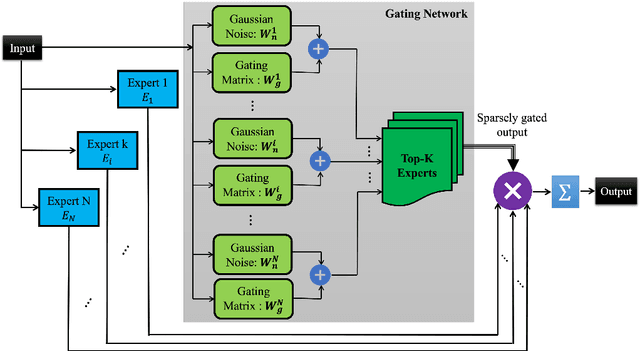
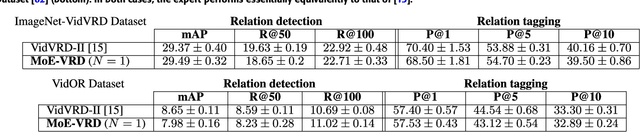
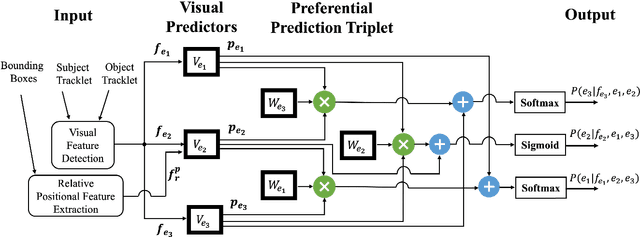
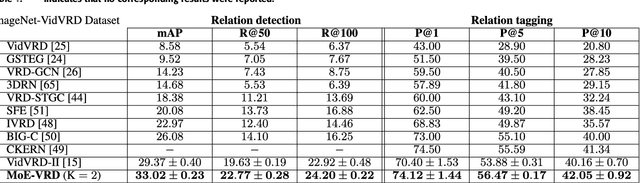
Abstract:Machine comprehension of visual information from images and videos by neural networks faces two primary challenges. Firstly, there exists a computational and inference gap in connecting vision and language, making it difficult to accurately determine which object a given agent acts on and represent it through language. Secondly, classifiers trained by a single, monolithic neural network often lack stability and generalization. To overcome these challenges, we introduce MoE-VRD, a novel approach to visual relationship detection utilizing a mixture of experts. MoE-VRD identifies language triplets in the form of < subject, predicate, object> tuples to extract relationships from visual processing. Leveraging recent advancements in visual relationship detection, MoE-VRD addresses the requirement for action recognition in establishing relationships between subjects (acting) and objects (being acted upon). In contrast to single monolithic networks, MoE-VRD employs multiple small models as experts, whose outputs are aggregated. Each expert in MoE-VRD specializes in visual relationship learning and object tagging. By utilizing a sparsely-gated mixture of experts, MoE-VRD enables conditional computation and significantly enhances neural network capacity without increasing computational complexity. Our experimental results demonstrate that the conditional computation capabilities and scalability of the mixture-of-experts approach lead to superior performance in visual relationship detection compared to state-of-the-art methods.
EPNet: An Efficient Pyramid Network for Enhanced Single-Image Super-Resolution with Reduced Computational Requirements
Dec 20, 2023Abstract:Single-image super-resolution (SISR) has seen significant advancements through the integration of deep learning. However, the substantial computational and memory requirements of existing methods often limit their practical application. This paper introduces a new Efficient Pyramid Network (EPNet) that harmoniously merges an Edge Split Pyramid Module (ESPM) with a Panoramic Feature Extraction Module (PFEM) to overcome the limitations of existing methods, particularly in terms of computational efficiency. The ESPM applies a pyramid-based channel separation strategy, boosting feature extraction while maintaining computational efficiency. The PFEM, a novel fusion of CNN and Transformer structures, enables the concurrent extraction of local and global features, thereby providing a panoramic view of the image landscape. Our architecture integrates the PFEM in a manner that facilitates the streamlined exchange of feature information and allows for the further refinement of image texture details. Experimental results indicate that our model outperforms existing state-of-the-art methods in image resolution quality, while considerably decreasing computational and memory costs. This research contributes to the ongoing evolution of efficient and practical SISR methodologies, bearing broader implications for the field of computer vision.
Challenges for Predictive Modeling with Neural Network Techniques using Error-Prone Dietary Intake Data
Nov 15, 2023



Abstract:Dietary intake data are routinely drawn upon to explore diet-health relationships. However, these data are often subject to measurement error, distorting the true relationships. Beyond measurement error, there are likely complex synergistic and sometimes antagonistic interactions between different dietary components, complicating the relationships between diet and health outcomes. Flexible models are required to capture the nuance that these complex interactions introduce. This complexity makes research on diet-health relationships an appealing candidate for the application of machine learning techniques, and in particular, neural networks. Neural networks are computational models that are able to capture highly complex, nonlinear relationships so long as sufficient data are available. While these models have been applied in many domains, the impacts of measurement error on the performance of predictive modeling has not been systematically investigated. However, dietary intake data are typically collected using self-report methods and are prone to large amounts of measurement error. In this work, we demonstrate the ways in which measurement error erodes the performance of neural networks, and illustrate the care that is required for leveraging these models in the presence of error. We demonstrate the role that sample size and replicate measurements play on model performance, indicate a motivation for the investigation of transformations to additivity, and illustrate the caution required to prevent model overfitting. While the past performance of neural networks across various domains make them an attractive candidate for examining diet-health relationships, our work demonstrates that substantial care and further methodological development are both required to observe increased predictive performance when applying these techniques, compared to more traditional statistical procedures.
Memory-Efficient Continual Learning Object Segmentation for Long Video
Sep 26, 2023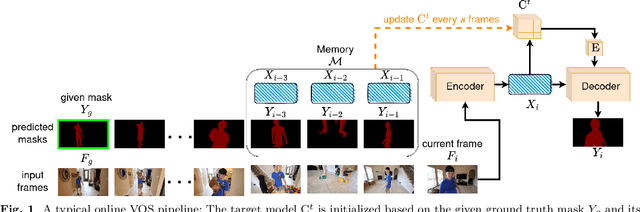



Abstract:Recent state-of-the-art semi-supervised Video Object Segmentation (VOS) methods have shown significant improvements in target object segmentation accuracy when information from preceding frames is used in undertaking segmentation on the current frame. In particular, such memory-based approaches can help a model to more effectively handle appearance changes (representation drift) or occlusions. Ideally, for maximum performance, online VOS methods would need all or most of the preceding frames (or their extracted information) to be stored in memory and be used for online learning in consecutive frames. Such a solution is not feasible for long videos, as the required memory size would grow without bound. On the other hand, these methods can fail when memory is limited and a target object experiences repeated representation drifts throughout a video. We propose two novel techniques to reduce the memory requirement of online VOS methods while improving modeling accuracy and generalization on long videos. Motivated by the success of continual learning techniques in preserving previously-learned knowledge, here we propose Gated-Regularizer Continual Learning (GRCL), which improves the performance of any online VOS subject to limited memory, and a Reconstruction-based Memory Selection Continual Learning (RMSCL) which empowers online VOS methods to efficiently benefit from stored information in memory. Experimental results show that the proposed methods improve the performance of online VOS models up to 10 %, and boosts their robustness on long-video datasets while maintaining comparable performance on short-video datasets DAVIS16 and DAVIS17.
A Step Towards Worldwide Biodiversity Assessment: The BIOSCAN-1M Insect Dataset
Jul 19, 2023Abstract:In an effort to catalog insect biodiversity, we propose a new large dataset of hand-labelled insect images, the BIOSCAN-Insect Dataset. Each record is taxonomically classified by an expert, and also has associated genetic information including raw nucleotide barcode sequences and assigned barcode index numbers, which are genetically-based proxies for species classification. This paper presents a curated million-image dataset, primarily to train computer-vision models capable of providing image-based taxonomic assessment, however, the dataset also presents compelling characteristics, the study of which would be of interest to the broader machine learning community. Driven by the biological nature inherent to the dataset, a characteristic long-tailed class-imbalance distribution is exhibited. Furthermore, taxonomic labelling is a hierarchical classification scheme, presenting a highly fine-grained classification problem at lower levels. Beyond spurring interest in biodiversity research within the machine learning community, progress on creating an image-based taxonomic classifier will also further the ultimate goal of all BIOSCAN research: to lay the foundation for a comprehensive survey of global biodiversity. This paper introduces the dataset and explores the classification task through the implementation and analysis of a baseline classifier.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge