Scott C. Lowe
A continental-scale dataset of ground beetles with high-resolution images and validated morphological trait measurements
Jan 14, 2026Abstract:Despite the ecological significance of invertebrates, global trait databases remain heavily biased toward vertebrates and plants, limiting comprehensive ecological analyses of high-diversity groups like ground beetles. Ground beetles (Coleoptera: Carabidae) serve as critical bioindicators of ecosystem health, providing valuable insights into biodiversity shifts driven by environmental changes. While the National Ecological Observatory Network (NEON) maintains an extensive collection of carabid specimens from across the United States, these primarily exist as physical collections, restricting widespread research access and large-scale analysis. To address these gaps, we present a multimodal dataset digitizing over 13,200 NEON carabids from 30 sites spanning the continental US and Hawaii through high-resolution imaging, enabling broader access and computational analysis. The dataset includes digitally measured elytra length and width of each specimen, establishing a foundation for automated trait extraction using AI. Validated against manual measurements, our digital trait extraction achieves sub-millimeter precision, ensuring reliability for ecological and computational studies. By addressing invertebrate under-representation in trait databases, this work supports AI-driven tools for automated species identification and trait-based research, fostering advancements in biodiversity monitoring and conservation.
BarcodeMamba+: Advancing State-Space Models for Fungal Biodiversity Research
Dec 17, 2025Abstract:Accurate taxonomic classification from DNA barcodes is a cornerstone of global biodiversity monitoring, yet fungi present extreme challenges due to sparse labelling and long-tailed taxa distributions. Conventional supervised learning methods often falter in this domain, struggling to generalize to unseen species and to capture the hierarchical nature of the data. To address these limitations, we introduce BarcodeMamba+, a foundation model for fungal barcode classification built on a powerful and efficient state-space model architecture. We employ a pretrain and fine-tune paradigm, which utilizes partially labelled data and we demonstrate this is substantially more effective than traditional fully-supervised methods in this data-sparse environment. During fine-tuning, we systematically integrate and evaluate a suite of enhancements--including hierarchical label smoothing, a weighted loss function, and a multi-head output layer from MycoAI--to specifically tackle the challenges of fungal taxonomy. Our experiments show that each of these components yields significant performance gains. On a challenging fungal classification benchmark with distinct taxonomic distribution shifts from the broad training set, our final model outperforms a range of existing methods across all taxonomic levels. Our work provides a powerful new tool for genomics-based biodiversity research and establishes an effective and scalable training paradigm for this challenging domain. Our code is publicly available at https://github.com/bioscan-ml/BarcodeMamba.
Understanding and Improving the Shampoo Optimizer via Kullback-Leibler Minimization
Sep 03, 2025Abstract:As an adaptive method, Shampoo employs a structured second-moment estimation, and its effectiveness has attracted growing attention. Prior work has primarily analyzed its estimation scheme through the Frobenius norm. Motivated by the natural connection between the second moment and a covariance matrix, we propose studying Shampoo's estimation as covariance estimation through the lens of Kullback-Leibler (KL) minimization. This alternative perspective reveals a previously hidden limitation, motivating improvements to Shampoo's design. Building on this insight, we develop a practical estimation scheme, termed KL-Shampoo, that eliminates Shampoo's reliance on Adam for stabilization, thereby removing the additional memory overhead introduced by Adam. Preliminary results show that KL-Shampoo improves Shampoo's performance, enabling it to stabilize without Adam and even outperform its Adam-stabilized variant, SOAP, in neural network pretraining.
A multi-modal dataset for insect biodiversity with imagery and DNA at the trap and individual level
Jul 09, 2025Abstract:Insects comprise millions of species, many experiencing severe population declines under environmental and habitat changes. High-throughput approaches are crucial for accelerating our understanding of insect diversity, with DNA barcoding and high-resolution imaging showing strong potential for automatic taxonomic classification. However, most image-based approaches rely on individual specimen data, unlike the unsorted bulk samples collected in large-scale ecological surveys. We present the Mixed Arthropod Sample Segmentation and Identification (MassID45) dataset for training automatic classifiers of bulk insect samples. It uniquely combines molecular and imaging data at both the unsorted sample level and the full set of individual specimens. Human annotators, supported by an AI-assisted tool, performed two tasks on bulk images: creating segmentation masks around each individual arthropod and assigning taxonomic labels to over 17 000 specimens. Combining the taxonomic resolution of DNA barcodes with precise abundance estimates of bulk images holds great potential for rapid, large-scale characterization of insect communities. This dataset pushes the boundaries of tiny object detection and instance segmentation, fostering innovation in both ecological and machine learning research.
Enhancing DNA Foundation Models to Address Masking Inefficiencies
Feb 25, 2025Abstract:Masked language modelling (MLM) as a pretraining objective has been widely adopted in genomic sequence modelling. While pretrained models can successfully serve as encoders for various downstream tasks, the distribution shift between pretraining and inference detrimentally impacts performance, as the pretraining task is to map [MASK] tokens to predictions, yet the [MASK] is absent during downstream applications. This means the encoder does not prioritize its encodings of non-[MASK] tokens, and expends parameters and compute on work only relevant to the MLM task, despite this being irrelevant at deployment time. In this work, we propose a modified encoder-decoder architecture based on the masked autoencoder framework, designed to address this inefficiency within a BERT-based transformer. We empirically show that the resulting mismatch is particularly detrimental in genomic pipelines where models are often used for feature extraction without fine-tuning. We evaluate our approach on the BIOSCAN-5M dataset, comprising over 2 million unique DNA barcodes. We achieve substantial performance gains in both closed-world and open-world classification tasks when compared against causal models and bidirectional architectures pretrained with MLM tasks.
System 2 reasoning capabilities are nigh
Oct 04, 2024Abstract:In recent years, machine learning models have made strides towards human-like reasoning capabilities from several directions. In this work, we review the current state of the literature and describe the remaining steps to achieve a neural model which can perform System 2 reasoning analogous to a human. We argue that if current models are insufficient to be classed as performing reasoning, there remains very little additional progress needed to attain that goal.
Hierarchical Multi-Label Classification with Missing Information for Benthic Habitat Imagery
Sep 10, 2024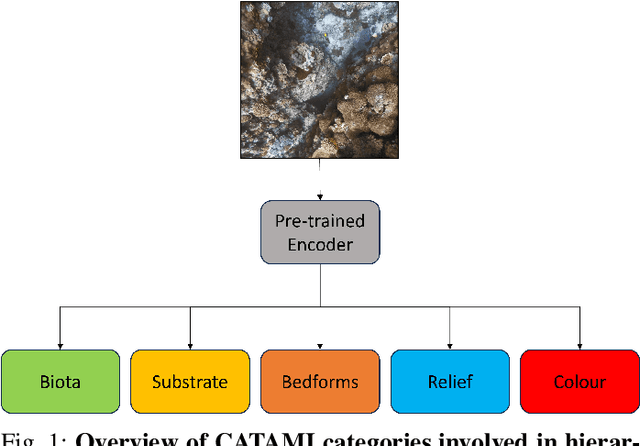
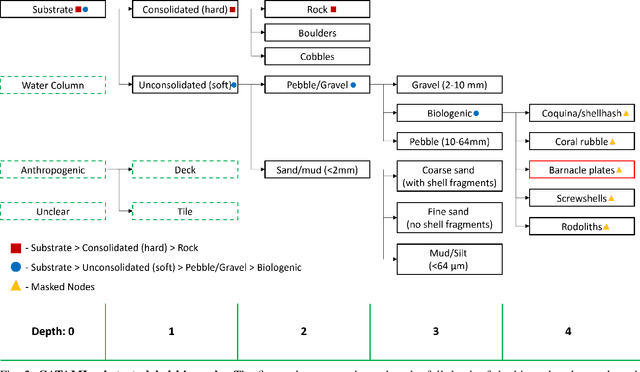
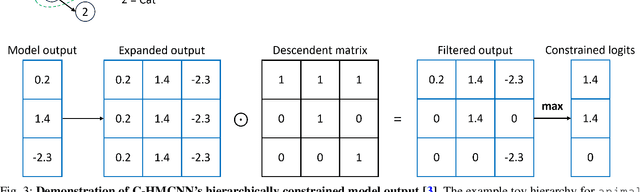
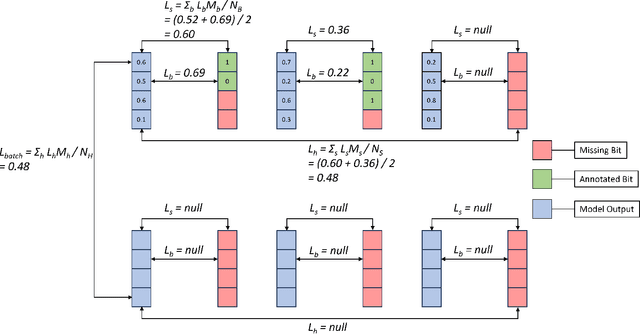
Abstract:In this work, we apply state-of-the-art self-supervised learning techniques on a large dataset of seafloor imagery, \textit{BenthicNet}, and study their performance for a complex hierarchical multi-label (HML) classification downstream task. In particular, we demonstrate the capacity to conduct HML training in scenarios where there exist multiple levels of missing annotation information, an important scenario for handling heterogeneous real-world data collected by multiple research groups with differing data collection protocols. We find that, when using smaller one-hot image label datasets typical of local or regional scale benthic science projects, models pre-trained with self-supervision on a larger collection of in-domain benthic data outperform models pre-trained on ImageNet. In the HML setting, we find the model can attain a deeper and more precise classification if it is pre-trained with self-supervision on in-domain data. We hope this work can establish a benchmark for future models in the field of automated underwater image annotation tasks and can guide work in other domains with hierarchical annotations of mixed resolution.
BIOSCAN-5M: A Multimodal Dataset for Insect Biodiversity
Jun 18, 2024



Abstract:As part of an ongoing worldwide effort to comprehend and monitor insect biodiversity, this paper presents the BIOSCAN-5M Insect dataset to the machine learning community and establish several benchmark tasks. BIOSCAN-5M is a comprehensive dataset containing multi-modal information for over 5 million insect specimens, and it significantly expands existing image-based biological datasets by including taxonomic labels, raw nucleotide barcode sequences, assigned barcode index numbers, and geographical information. We propose three benchmark experiments to demonstrate the impact of the multi-modal data types on the classification and clustering accuracy. First, we pretrain a masked language model on the DNA barcode sequences of the \mbox{BIOSCAN-5M} dataset, and demonstrate the impact of using this large reference library on species- and genus-level classification performance. Second, we propose a zero-shot transfer learning task applied to images and DNA barcodes to cluster feature embeddings obtained from self-supervised learning, to investigate whether meaningful clusters can be derived from these representation embeddings. Third, we benchmark multi-modality by performing contrastive learning on DNA barcodes, image data, and taxonomic information. This yields a general shared embedding space enabling taxonomic classification using multiple types of information and modalities. The code repository of the BIOSCAN-5M Insect dataset is available at {\url{https://github.com/zahrag/BIOSCAN-5M}}
An Empirical Study into Clustering of Unseen Datasets with Self-Supervised Encoders
Jun 04, 2024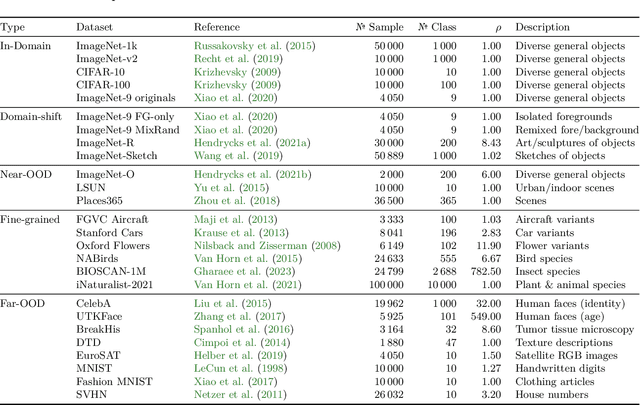
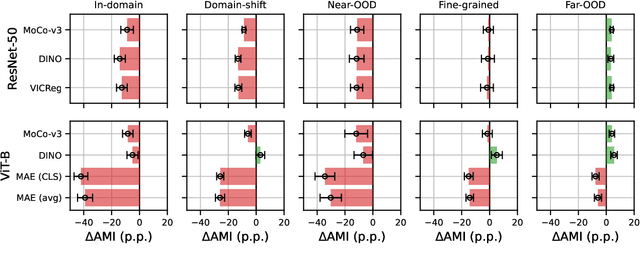
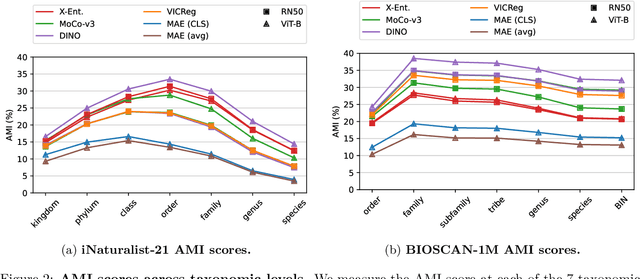
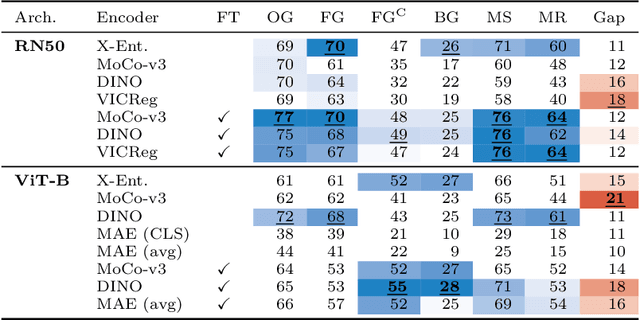
Abstract:Can pretrained models generalize to new datasets without any retraining? We deploy pretrained image models on datasets they were not trained for, and investigate whether their embeddings form meaningful clusters. Our suite of benchmarking experiments use encoders pretrained solely on ImageNet-1k with either supervised or self-supervised training techniques, deployed on image datasets that were not seen during training, and clustered with conventional clustering algorithms. This evaluation provides new insights into the embeddings of self-supervised models, which prioritize different features to supervised models. Supervised encoders typically offer more utility than SSL encoders within the training domain, and vice-versa far outside of it, however, fine-tuned encoders demonstrate the opposite trend. Clustering provides a way to evaluate the utility of self-supervised learned representations orthogonal to existing methods such as kNN. Additionally, we find the silhouette score when measured in a UMAP-reduced space is highly correlated with clustering performance, and can therefore be used as a proxy for clustering performance on data with no ground truth labels. Our code implementation is available at \url{https://github.com/scottclowe/zs-ssl-clustering/}.
BIOSCAN-CLIP: Bridging Vision and Genomics for Biodiversity Monitoring at Scale
May 27, 2024

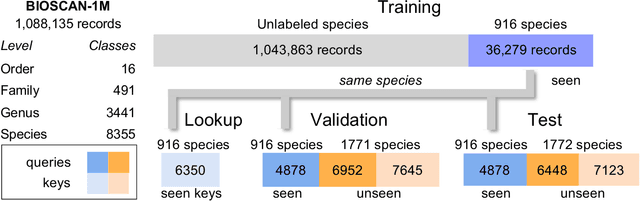

Abstract:Measuring biodiversity is crucial for understanding ecosystem health. While prior works have developed machine learning models for the taxonomic classification of photographic images and DNA separately, in this work, we introduce a multimodal approach combining both, using CLIP-style contrastive learning to align images, DNA barcodes, and textual data in a unified embedding space. This allows for accurate classification of both known and unknown insect species without task-specific fine-tuning, leveraging contrastive learning for the first time to fuse DNA and image data. Our method surpasses previous single-modality approaches in accuracy by over 11% on zero-shot learning tasks, showcasing its effectiveness in biodiversity studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge