Isaac Xu
Improving Detection of Rare Nodes in Hierarchical Multi-Label Learning
Feb 09, 2026Abstract:In hierarchical multi-label classification, a persistent challenge is enabling model predictions to reach deeper levels of the hierarchy for more detailed or fine-grained classifications. This difficulty partly arises from the natural rarity of certain classes (or hierarchical nodes) and the hierarchical constraint that ensures child nodes are almost always less frequent than their parents. To address this, we propose a weighted loss objective for neural networks that combines node-wise imbalance weighting with focal weighting components, the latter leveraging modern quantification of ensemble uncertainties. By emphasizing rare nodes rather than rare observations (data points), and focusing on uncertain nodes for each model output distribution during training, we observe improvements in recall by up to a factor of five on benchmark datasets, along with statistically significant gains in $F_{1}$ score. We also show our approach aids convolutional networks on challenging tasks, as in situations with suboptimal encoders or limited data.
Variance-Gated Ensembles: An Epistemic-Aware Framework for Uncertainty Estimation
Feb 08, 2026Abstract:Machine learning applications require fast and reliable per-sample uncertainty estimation. A common approach is to use predictive distributions from Bayesian or approximation methods and additively decompose uncertainty into aleatoric (i.e., data-related) and epistemic (i.e., model-related) components. However, additive decomposition has recently been questioned, with evidence that it breaks down when using finite-ensemble sampling and/or mismatched predictive distributions. This paper introduces Variance-Gated Ensembles (VGE), an intuitive, differentiable framework that injects epistemic sensitivity via a signal-to-noise gate computed from ensemble statistics. VGE provides: (i) a Variance-Gated Margin Uncertainty (VGMU) score that couples decision margins with ensemble predictive variance; and (ii) a Variance-Gated Normalization (VGN) layer that generalizes the variance-gated uncertainty mechanism to training via per-class, learnable normalization of ensemble member probabilities. We derive closed-form vector-Jacobian products enabling end-to-end training through ensemble sample mean and variance. VGE matches or exceeds state-of-the-art information-theoretic baselines while remaining computationally efficient. As a result, VGE provides a practical and scalable approach to epistemic-aware uncertainty estimation in ensemble models. An open-source implementation is available at: https://github.com/nextdevai/vge.
Label-free Monitoring of Self-Supervised Learning Progress
Sep 10, 2024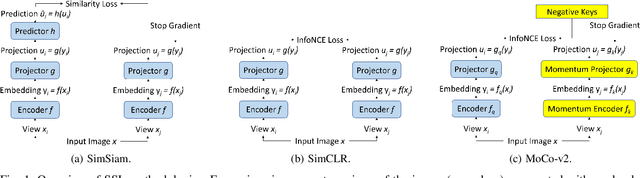
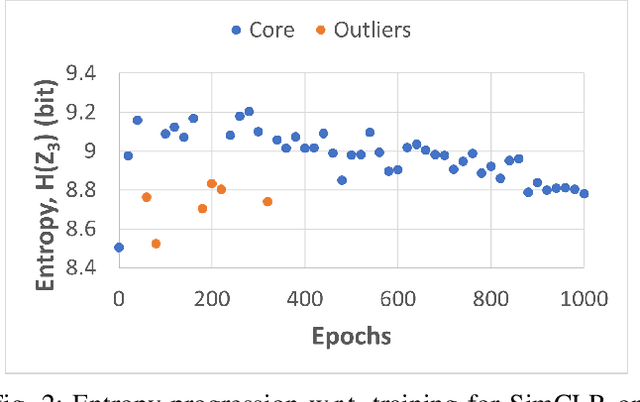
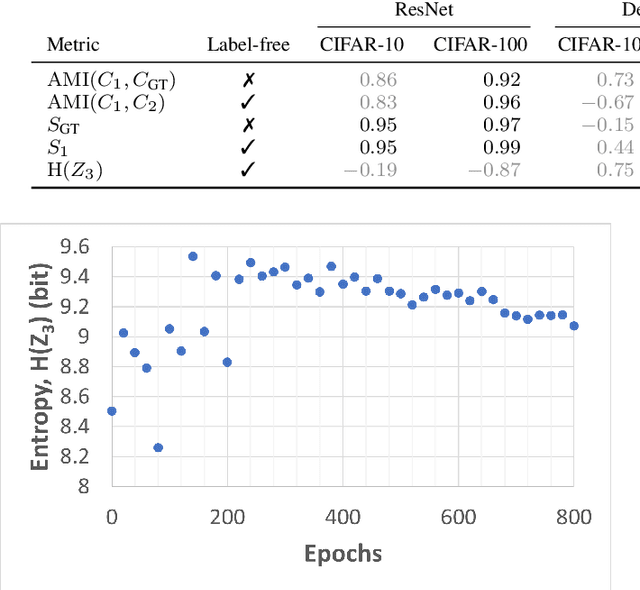
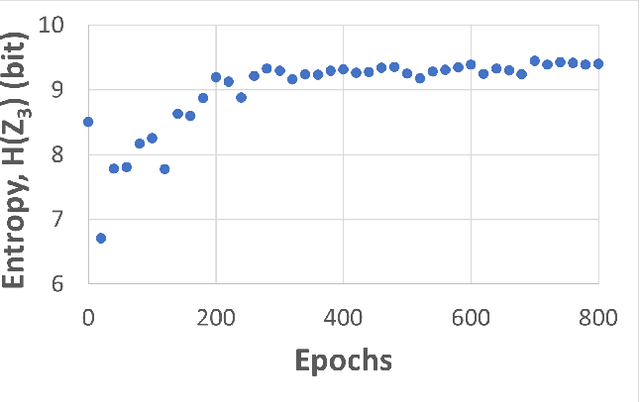
Abstract:Self-supervised learning (SSL) is an effective method for exploiting unlabelled data to learn a high-level embedding space that can be used for various downstream tasks. However, existing methods to monitor the quality of the encoder -- either during training for one model or to compare several trained models -- still rely on access to annotated data. When SSL methodologies are applied to new data domains, a sufficiently large labelled dataset may not always be available. In this study, we propose several evaluation metrics which can be applied on the embeddings of unlabelled data and investigate their viability by comparing them to linear probe accuracy (a common metric which utilizes an annotated dataset). In particular, we apply $k$-means clustering and measure the clustering quality with the silhouette score and clustering agreement. We also measure the entropy of the embedding distribution. We find that while the clusters did correspond better to the ground truth annotations as training of the network progressed, label-free clustering metrics correlated with the linear probe accuracy only when training with SSL methods SimCLR and MoCo-v2, but not with SimSiam. Additionally, although entropy did not always have strong correlations with LP accuracy, this appears to be due to instability arising from early training, with the metric stabilizing and becoming more reliable at later stages of learning. Furthermore, while entropy generally decreases as learning progresses, this trend reverses for SimSiam. More research is required to establish the cause for this unexpected behaviour. Lastly, we find that while clustering based approaches are likely only viable for same-architecture comparisons, entropy may be architecture-independent.
Hierarchical Multi-Label Classification with Missing Information for Benthic Habitat Imagery
Sep 10, 2024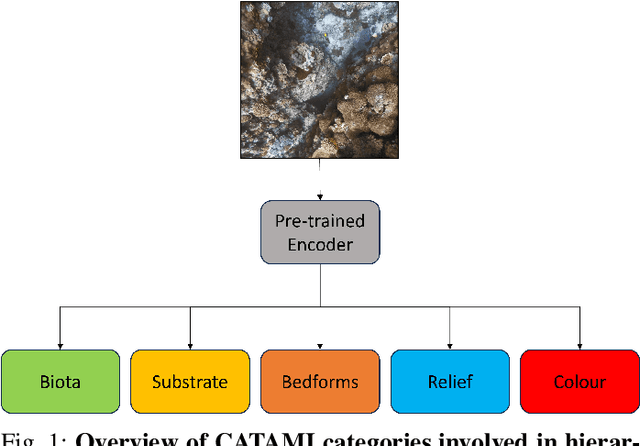
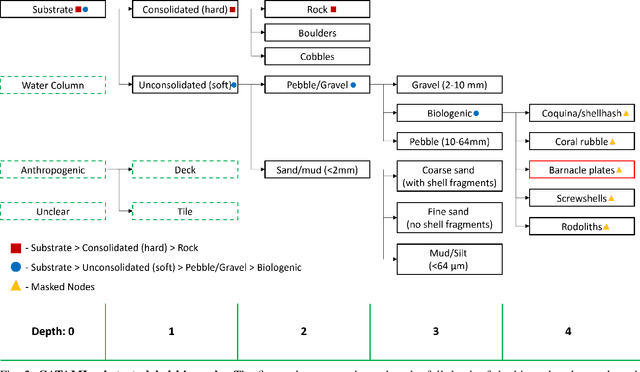
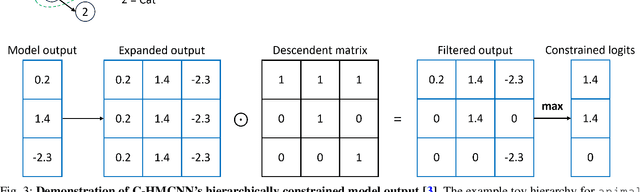
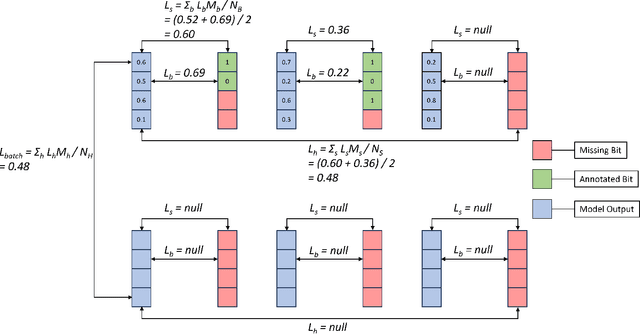
Abstract:In this work, we apply state-of-the-art self-supervised learning techniques on a large dataset of seafloor imagery, \textit{BenthicNet}, and study their performance for a complex hierarchical multi-label (HML) classification downstream task. In particular, we demonstrate the capacity to conduct HML training in scenarios where there exist multiple levels of missing annotation information, an important scenario for handling heterogeneous real-world data collected by multiple research groups with differing data collection protocols. We find that, when using smaller one-hot image label datasets typical of local or regional scale benthic science projects, models pre-trained with self-supervision on a larger collection of in-domain benthic data outperform models pre-trained on ImageNet. In the HML setting, we find the model can attain a deeper and more precise classification if it is pre-trained with self-supervision on in-domain data. We hope this work can establish a benchmark for future models in the field of automated underwater image annotation tasks and can guide work in other domains with hierarchical annotations of mixed resolution.
BenthicNet: A global compilation of seafloor images for deep learning applications
May 08, 2024Abstract:Advances in underwater imaging enable the collection of extensive seafloor image datasets that are necessary for monitoring important benthic ecosystems. The ability to collect seafloor imagery has outpaced our capacity to analyze it, hindering expedient mobilization of this crucial environmental information. Recent machine learning approaches provide opportunities to increase the efficiency with which seafloor image datasets are analyzed, yet large and consistent datasets necessary to support development of such approaches are scarce. Here we present BenthicNet: a global compilation of seafloor imagery designed to support the training and evaluation of large-scale image recognition models. An initial set of over 11.4 million images was collected and curated to represent a diversity of seafloor environments using a representative subset of 1.3 million images. These are accompanied by 2.6 million annotations translated to the CATAMI scheme, which span 190,000 of the images. A large deep learning model was trained on this compilation and preliminary results suggest it has utility for automating large and small-scale image analysis tasks. The compilation and model are made openly available for use by the scientific community at https://doi.org/10.20383/103.0614.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge