ZeMing Gong
ViGiL3D: A Linguistically Diverse Dataset for 3D Visual Grounding
Jan 02, 2025



Abstract:3D visual grounding (3DVG) involves localizing entities in a 3D scene referred to by natural language text. Such models are useful for embodied AI and scene retrieval applications, which involve searching for objects or patterns using natural language descriptions. While recent works have focused on LLM-based scaling of 3DVG datasets, these datasets do not capture the full range of potential prompts which could be specified in the English language. To ensure that we are scaling up and testing against a useful and representative set of prompts, we propose a framework for linguistically analyzing 3DVG prompts and introduce Visual Grounding with Diverse Language in 3D (ViGiL3D), a diagnostic dataset for evaluating visual grounding methods against a diverse set of language patterns. We evaluate existing open-vocabulary 3DVG methods to demonstrate that these methods are not yet proficient in understanding and identifying the targets of more challenging, out-of-distribution prompts, toward real-world applications.
BIOSCAN-5M: A Multimodal Dataset for Insect Biodiversity
Jun 18, 2024



Abstract:As part of an ongoing worldwide effort to comprehend and monitor insect biodiversity, this paper presents the BIOSCAN-5M Insect dataset to the machine learning community and establish several benchmark tasks. BIOSCAN-5M is a comprehensive dataset containing multi-modal information for over 5 million insect specimens, and it significantly expands existing image-based biological datasets by including taxonomic labels, raw nucleotide barcode sequences, assigned barcode index numbers, and geographical information. We propose three benchmark experiments to demonstrate the impact of the multi-modal data types on the classification and clustering accuracy. First, we pretrain a masked language model on the DNA barcode sequences of the \mbox{BIOSCAN-5M} dataset, and demonstrate the impact of using this large reference library on species- and genus-level classification performance. Second, we propose a zero-shot transfer learning task applied to images and DNA barcodes to cluster feature embeddings obtained from self-supervised learning, to investigate whether meaningful clusters can be derived from these representation embeddings. Third, we benchmark multi-modality by performing contrastive learning on DNA barcodes, image data, and taxonomic information. This yields a general shared embedding space enabling taxonomic classification using multiple types of information and modalities. The code repository of the BIOSCAN-5M Insect dataset is available at {\url{https://github.com/zahrag/BIOSCAN-5M}}
BIOSCAN-CLIP: Bridging Vision and Genomics for Biodiversity Monitoring at Scale
May 27, 2024


Abstract:Measuring biodiversity is crucial for understanding ecosystem health. While prior works have developed machine learning models for the taxonomic classification of photographic images and DNA separately, in this work, we introduce a multimodal approach combining both, using CLIP-style contrastive learning to align images, DNA barcodes, and textual data in a unified embedding space. This allows for accurate classification of both known and unknown insect species without task-specific fine-tuning, leveraging contrastive learning for the first time to fuse DNA and image data. Our method surpasses previous single-modality approaches in accuracy by over 11% on zero-shot learning tasks, showcasing its effectiveness in biodiversity studies.
BarcodeBERT: Transformers for Biodiversity Analysis
Nov 04, 2023Abstract:Understanding biodiversity is a global challenge, in which DNA barcodes - short snippets of DNA that cluster by species - play a pivotal role. In particular, invertebrates, a highly diverse and under-explored group, pose unique taxonomic complexities. We explore machine learning approaches, comparing supervised CNNs, fine-tuned foundation models, and a DNA barcode-specific masking strategy across datasets of varying complexity. While simpler datasets and tasks favor supervised CNNs or fine-tuned transformers, challenging species-level identification demands a paradigm shift towards self-supervised pretraining. We propose BarcodeBERT, the first self-supervised method for general biodiversity analysis, leveraging a 1.5 M invertebrate DNA barcode reference library. This work highlights how dataset specifics and coverage impact model selection, and underscores the role of self-supervised pretraining in achieving high-accuracy DNA barcode-based identification at the species and genus level. Indeed, without the fine-tuning step, BarcodeBERT pretrained on a large DNA barcode dataset outperforms DNABERT and DNABERT-2 on multiple downstream classification tasks. The code repository is available at https://github.com/Kari-Genomics-Lab/BarcodeBERT
Multi3DRefer: Grounding Text Description to Multiple 3D Objects
Sep 11, 2023Abstract:We introduce the task of localizing a flexible number of objects in real-world 3D scenes using natural language descriptions. Existing 3D visual grounding tasks focus on localizing a unique object given a text description. However, such a strict setting is unnatural as localizing potentially multiple objects is a common need in real-world scenarios and robotic tasks (e.g., visual navigation and object rearrangement). To address this setting we propose Multi3DRefer, generalizing the ScanRefer dataset and task. Our dataset contains 61926 descriptions of 11609 objects, where zero, single or multiple target objects are referenced by each description. We also introduce a new evaluation metric and benchmark methods from prior work to enable further investigation of multi-modal 3D scene understanding. Furthermore, we develop a better baseline leveraging 2D features from CLIP by rendering object proposals online with contrastive learning, which outperforms the state of the art on the ScanRefer benchmark.
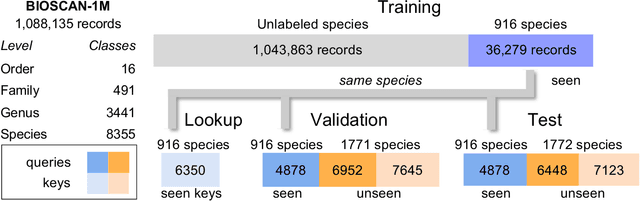
A Step Towards Worldwide Biodiversity Assessment: The BIOSCAN-1M Insect Dataset
Jul 19, 2023Abstract:In an effort to catalog insect biodiversity, we propose a new large dataset of hand-labelled insect images, the BIOSCAN-Insect Dataset. Each record is taxonomically classified by an expert, and also has associated genetic information including raw nucleotide barcode sequences and assigned barcode index numbers, which are genetically-based proxies for species classification. This paper presents a curated million-image dataset, primarily to train computer-vision models capable of providing image-based taxonomic assessment, however, the dataset also presents compelling characteristics, the study of which would be of interest to the broader machine learning community. Driven by the biological nature inherent to the dataset, a characteristic long-tailed class-imbalance distribution is exhibited. Furthermore, taxonomic labelling is a hierarchical classification scheme, presenting a highly fine-grained classification problem at lower levels. Beyond spurring interest in biodiversity research within the machine learning community, progress on creating an image-based taxonomic classifier will also further the ultimate goal of all BIOSCAN research: to lay the foundation for a comprehensive survey of global biodiversity. This paper introduces the dataset and explores the classification task through the implementation and analysis of a baseline classifier.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge