Austin T. Wang
HSM: Hierarchical Scene Motifs for Multi-Scale Indoor Scene Generation
Mar 21, 2025
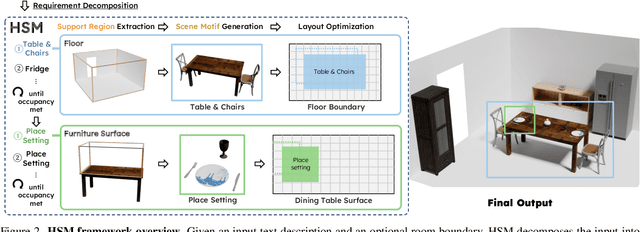

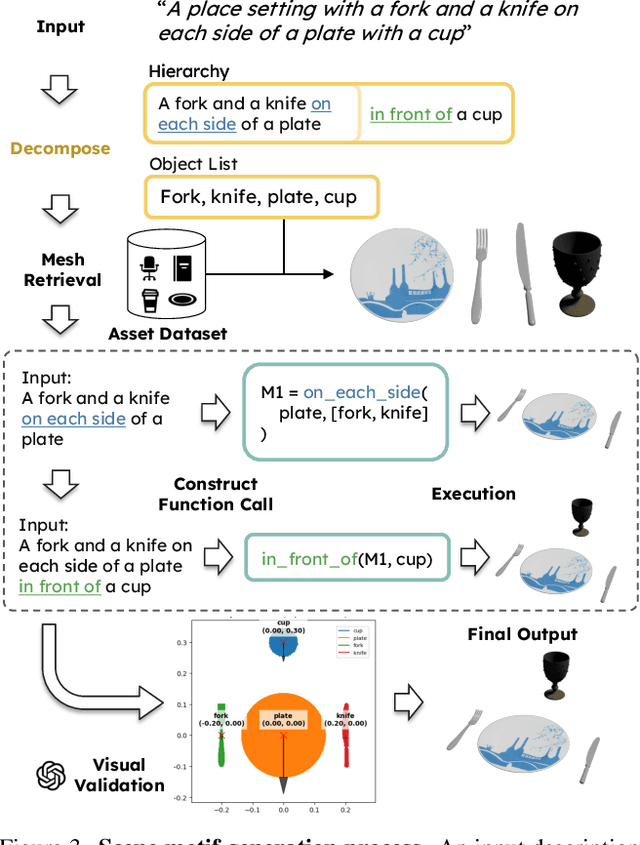
Abstract:Despite advances in indoor 3D scene layout generation, synthesizing scenes with dense object arrangements remains challenging. Existing methods primarily focus on large furniture while neglecting smaller objects, resulting in unrealistically empty scenes. Those that place small objects typically do not honor arrangement specifications, resulting in largely random placement not following the text description. We present HSM, a hierarchical framework for indoor scene generation with dense object arrangements across spatial scales. Indoor scenes are inherently hierarchical, with surfaces supporting objects at different scales, from large furniture on floors to smaller objects on tables and shelves. HSM embraces this hierarchy and exploits recurring cross-scale spatial patterns to generate complex and realistic indoor scenes in a unified manner. Our experiments show that HSM outperforms existing methods by generating scenes that are more realistic and better conform to user input across room types and spatial configurations.
SceneEval: Evaluating Semantic Coherence in Text-Conditioned 3D Indoor Scene Synthesis
Mar 18, 2025
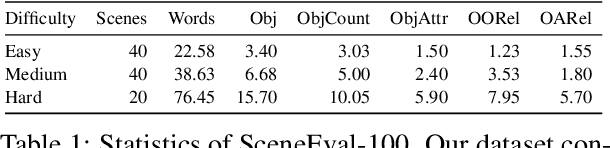
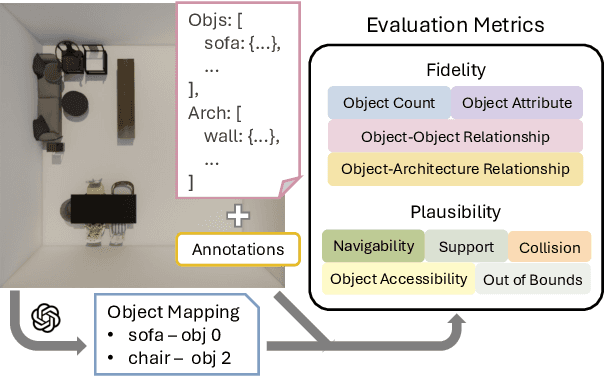

Abstract:Despite recent advances in text-conditioned 3D indoor scene generation, there remain gaps in the evaluation of these methods. Existing metrics primarily assess the realism of generated scenes by comparing them to a set of ground-truth scenes, often overlooking alignment with the input text - a critical factor in determining how effectively a method meets user requirements. We present SceneEval, an evaluation framework designed to address this limitation. SceneEval includes metrics for both explicit user requirements, such as the presence of specific objects and their attributes described in the input text, and implicit expectations, like the absence of object collisions, providing a comprehensive assessment of scene quality. To facilitate evaluation, we introduce SceneEval-100, a dataset of scene descriptions with annotated ground-truth scene properties. We evaluate recent scene generation methods using SceneEval and demonstrate its ability to provide detailed assessments of the generated scenes, highlighting strengths and areas for improvement across multiple dimensions. Our results show that current methods struggle at generating scenes that meet user requirements, underscoring the need for further research in this direction.
ViGiL3D: A Linguistically Diverse Dataset for 3D Visual Grounding
Jan 02, 2025



Abstract:3D visual grounding (3DVG) involves localizing entities in a 3D scene referred to by natural language text. Such models are useful for embodied AI and scene retrieval applications, which involve searching for objects or patterns using natural language descriptions. While recent works have focused on LLM-based scaling of 3DVG datasets, these datasets do not capture the full range of potential prompts which could be specified in the English language. To ensure that we are scaling up and testing against a useful and representative set of prompts, we propose a framework for linguistically analyzing 3DVG prompts and introduce Visual Grounding with Diverse Language in 3D (ViGiL3D), a diagnostic dataset for evaluating visual grounding methods against a diverse set of language patterns. We evaluate existing open-vocabulary 3DVG methods to demonstrate that these methods are not yet proficient in understanding and identifying the targets of more challenging, out-of-distribution prompts, toward real-world applications.
BIOSCAN-5M: A Multimodal Dataset for Insect Biodiversity
Jun 18, 2024



Abstract:As part of an ongoing worldwide effort to comprehend and monitor insect biodiversity, this paper presents the BIOSCAN-5M Insect dataset to the machine learning community and establish several benchmark tasks. BIOSCAN-5M is a comprehensive dataset containing multi-modal information for over 5 million insect specimens, and it significantly expands existing image-based biological datasets by including taxonomic labels, raw nucleotide barcode sequences, assigned barcode index numbers, and geographical information. We propose three benchmark experiments to demonstrate the impact of the multi-modal data types on the classification and clustering accuracy. First, we pretrain a masked language model on the DNA barcode sequences of the \mbox{BIOSCAN-5M} dataset, and demonstrate the impact of using this large reference library on species- and genus-level classification performance. Second, we propose a zero-shot transfer learning task applied to images and DNA barcodes to cluster feature embeddings obtained from self-supervised learning, to investigate whether meaningful clusters can be derived from these representation embeddings. Third, we benchmark multi-modality by performing contrastive learning on DNA barcodes, image data, and taxonomic information. This yields a general shared embedding space enabling taxonomic classification using multiple types of information and modalities. The code repository of the BIOSCAN-5M Insect dataset is available at {\url{https://github.com/zahrag/BIOSCAN-5M}}
BIOSCAN-CLIP: Bridging Vision and Genomics for Biodiversity Monitoring at Scale
May 27, 2024

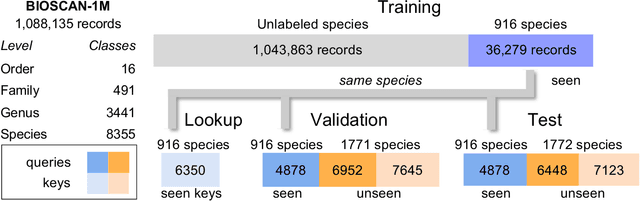

Abstract:Measuring biodiversity is crucial for understanding ecosystem health. While prior works have developed machine learning models for the taxonomic classification of photographic images and DNA separately, in this work, we introduce a multimodal approach combining both, using CLIP-style contrastive learning to align images, DNA barcodes, and textual data in a unified embedding space. This allows for accurate classification of both known and unknown insect species without task-specific fine-tuning, leveraging contrastive learning for the first time to fuse DNA and image data. Our method surpasses previous single-modality approaches in accuracy by over 11% on zero-shot learning tasks, showcasing its effectiveness in biodiversity studies.
BarcodeBERT: Transformers for Biodiversity Analysis
Nov 04, 2023Abstract:Understanding biodiversity is a global challenge, in which DNA barcodes - short snippets of DNA that cluster by species - play a pivotal role. In particular, invertebrates, a highly diverse and under-explored group, pose unique taxonomic complexities. We explore machine learning approaches, comparing supervised CNNs, fine-tuned foundation models, and a DNA barcode-specific masking strategy across datasets of varying complexity. While simpler datasets and tasks favor supervised CNNs or fine-tuned transformers, challenging species-level identification demands a paradigm shift towards self-supervised pretraining. We propose BarcodeBERT, the first self-supervised method for general biodiversity analysis, leveraging a 1.5 M invertebrate DNA barcode reference library. This work highlights how dataset specifics and coverage impact model selection, and underscores the role of self-supervised pretraining in achieving high-accuracy DNA barcode-based identification at the species and genus level. Indeed, without the fine-tuning step, BarcodeBERT pretrained on a large DNA barcode dataset outperforms DNABERT and DNABERT-2 on multiple downstream classification tasks. The code repository is available at https://github.com/Kari-Genomics-Lab/BarcodeBERT
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge