Joakim Bruslund Haurum
How to Sample High Quality 3D Fractals for Action Recognition Pre-Training?
Feb 12, 2026Abstract:Synthetic datasets are being recognized in the deep learning realm as a valuable alternative to exhaustively labeled real data. One such synthetic data generation method is Formula Driven Supervised Learning (FDSL), which can provide an infinite number of perfectly labeled data through a formula driven approach, such as fractals or contours. FDSL does not have common drawbacks like manual labor, privacy and other ethical concerns. In this work we generate 3D fractals using 3D Iterated Function Systems (IFS) for pre-training an action recognition model. The fractals are temporally transformed to form a video that is used as a pre-training dataset for downstream task of action recognition. We find that standard methods of generating fractals are slow and produce degenerate 3D fractals. Therefore, we systematically explore alternative ways of generating fractals and finds that overly-restrictive approaches, while generating aesthetically pleasing fractals, are detrimental for downstream task performance. We propose a novel method, Targeted Smart Filtering, to address both the generation speed and fractal diversity issue. The method reports roughly 100 times faster sampling speed and achieves superior downstream performance against other 3D fractal filtering methods.
Comparing Euclidean and Hyperbolic K-Means for Generalized Category Discovery
Feb 04, 2026Abstract:Hyperbolic representation learning has been widely used to extract implicit hierarchies within data, and recently it has found its way to the open-world classification task of Generalized Category Discovery (GCD). However, prior hyperbolic GCD methods only use hyperbolic geometry for representation learning and transform back to Euclidean geometry when clustering. We hypothesize this is suboptimal. Therefore, we present Hyperbolic Clustered GCD (HC-GCD), which learns embeddings in the Lorentz Hyperboloid model of hyperbolic geometry, and clusters these embeddings directly in hyperbolic space using a hyperbolic K-Means algorithm. We test our model on the Semantic Shift Benchmark datasets, and demonstrate that HC-GCD is on par with the previous state-of-the-art hyperbolic GCD method. Furthermore, we show that using hyperbolic K-Means leads to better accuracy than Euclidean K-Means. We carry out ablation studies showing that clipping the norm of the Euclidean embeddings leads to decreased accuracy in clustering unseen classes, and increased accuracy for seen classes, while the overall accuracy is dataset dependent. We also show that using hyperbolic K-Means leads to more consistent clusters when varying the label granularity.
Agglomerative Token Clustering
Sep 18, 2024Abstract:We present Agglomerative Token Clustering (ATC), a novel token merging method that consistently outperforms previous token merging and pruning methods across image classification, image synthesis, and object detection & segmentation tasks. ATC merges clusters through bottom-up hierarchical clustering, without the introduction of extra learnable parameters. We find that ATC achieves state-of-the-art performance across all tasks, and can even perform on par with prior state-of-the-art when applied off-the-shelf, i.e. without fine-tuning. ATC is particularly effective when applied with low keep rates, where only a small fraction of tokens are kept and retaining task performance is especially difficult.
BIOSCAN-5M: A Multimodal Dataset for Insect Biodiversity
Jun 18, 2024



Abstract:As part of an ongoing worldwide effort to comprehend and monitor insect biodiversity, this paper presents the BIOSCAN-5M Insect dataset to the machine learning community and establish several benchmark tasks. BIOSCAN-5M is a comprehensive dataset containing multi-modal information for over 5 million insect specimens, and it significantly expands existing image-based biological datasets by including taxonomic labels, raw nucleotide barcode sequences, assigned barcode index numbers, and geographical information. We propose three benchmark experiments to demonstrate the impact of the multi-modal data types on the classification and clustering accuracy. First, we pretrain a masked language model on the DNA barcode sequences of the \mbox{BIOSCAN-5M} dataset, and demonstrate the impact of using this large reference library on species- and genus-level classification performance. Second, we propose a zero-shot transfer learning task applied to images and DNA barcodes to cluster feature embeddings obtained from self-supervised learning, to investigate whether meaningful clusters can be derived from these representation embeddings. Third, we benchmark multi-modality by performing contrastive learning on DNA barcodes, image data, and taxonomic information. This yields a general shared embedding space enabling taxonomic classification using multiple types of information and modalities. The code repository of the BIOSCAN-5M Insect dataset is available at {\url{https://github.com/zahrag/BIOSCAN-5M}}
An Empirical Study into Clustering of Unseen Datasets with Self-Supervised Encoders
Jun 04, 2024
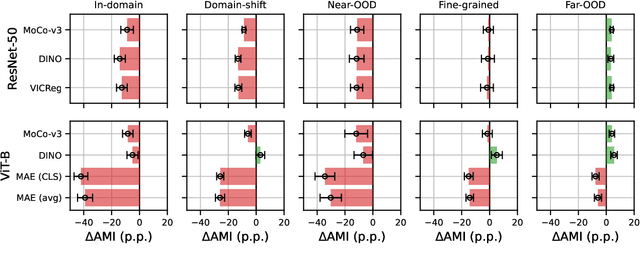
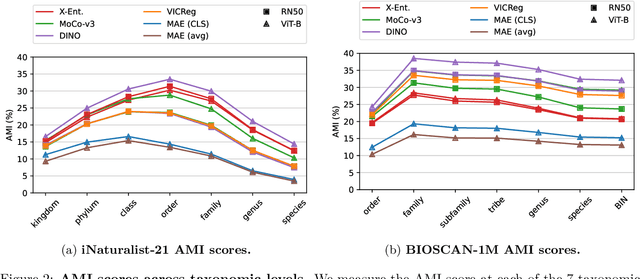
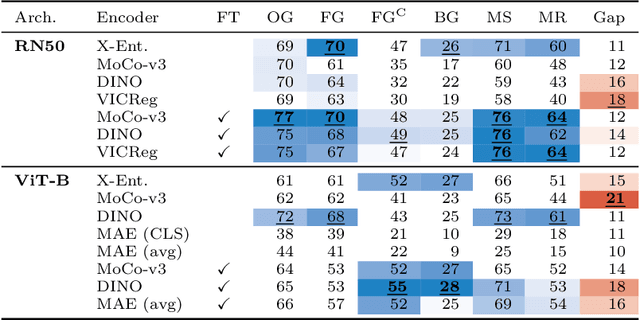
Abstract:Can pretrained models generalize to new datasets without any retraining? We deploy pretrained image models on datasets they were not trained for, and investigate whether their embeddings form meaningful clusters. Our suite of benchmarking experiments use encoders pretrained solely on ImageNet-1k with either supervised or self-supervised training techniques, deployed on image datasets that were not seen during training, and clustered with conventional clustering algorithms. This evaluation provides new insights into the embeddings of self-supervised models, which prioritize different features to supervised models. Supervised encoders typically offer more utility than SSL encoders within the training domain, and vice-versa far outside of it, however, fine-tuned encoders demonstrate the opposite trend. Clustering provides a way to evaluate the utility of self-supervised learned representations orthogonal to existing methods such as kNN. Additionally, we find the silhouette score when measured in a UMAP-reduced space is highly correlated with clustering performance, and can therefore be used as a proxy for clustering performance on data with no ground truth labels. Our code implementation is available at \url{https://github.com/scottclowe/zs-ssl-clustering/}.
BIOSCAN-CLIP: Bridging Vision and Genomics for Biodiversity Monitoring at Scale
May 27, 2024


Abstract:Measuring biodiversity is crucial for understanding ecosystem health. While prior works have developed machine learning models for the taxonomic classification of photographic images and DNA separately, in this work, we introduce a multimodal approach combining both, using CLIP-style contrastive learning to align images, DNA barcodes, and textual data in a unified embedding space. This allows for accurate classification of both known and unknown insect species without task-specific fine-tuning, leveraging contrastive learning for the first time to fuse DNA and image data. Our method surpasses previous single-modality approaches in accuracy by over 11% on zero-shot learning tasks, showcasing its effectiveness in biodiversity studies.
BarcodeBERT: Transformers for Biodiversity Analysis
Nov 04, 2023Abstract:Understanding biodiversity is a global challenge, in which DNA barcodes - short snippets of DNA that cluster by species - play a pivotal role. In particular, invertebrates, a highly diverse and under-explored group, pose unique taxonomic complexities. We explore machine learning approaches, comparing supervised CNNs, fine-tuned foundation models, and a DNA barcode-specific masking strategy across datasets of varying complexity. While simpler datasets and tasks favor supervised CNNs or fine-tuned transformers, challenging species-level identification demands a paradigm shift towards self-supervised pretraining. We propose BarcodeBERT, the first self-supervised method for general biodiversity analysis, leveraging a 1.5 M invertebrate DNA barcode reference library. This work highlights how dataset specifics and coverage impact model selection, and underscores the role of self-supervised pretraining in achieving high-accuracy DNA barcode-based identification at the species and genus level. Indeed, without the fine-tuning step, BarcodeBERT pretrained on a large DNA barcode dataset outperforms DNABERT and DNABERT-2 on multiple downstream classification tasks. The code repository is available at https://github.com/Kari-Genomics-Lab/BarcodeBERT
Which Tokens to Use? Investigating Token Reduction in Vision Transformers
Aug 09, 2023Abstract:Since the introduction of the Vision Transformer (ViT), researchers have sought to make ViTs more efficient by removing redundant information in the processed tokens. While different methods have been explored to achieve this goal, we still lack understanding of the resulting reduction patterns and how those patterns differ across token reduction methods and datasets. To close this gap, we set out to understand the reduction patterns of 10 different token reduction methods using four image classification datasets. By systematically comparing these methods on the different classification tasks, we find that the Top-K pruning method is a surprisingly strong baseline. Through in-depth analysis of the different methods, we determine that: the reduction patterns are generally not consistent when varying the capacity of the backbone model, the reduction patterns of pruning-based methods significantly differ from fixed radial patterns, and the reduction patterns of pruning-based methods are correlated across classification datasets. Finally we report that the similarity of reduction patterns is a moderate-to-strong proxy for model performance. Project page at https://vap.aau.dk/tokens.
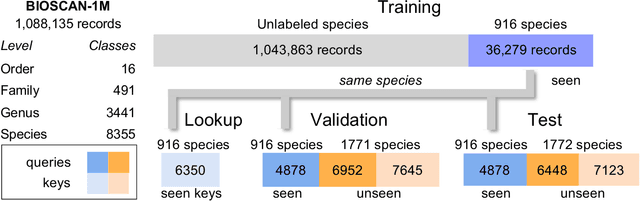
A Step Towards Worldwide Biodiversity Assessment: The BIOSCAN-1M Insect Dataset
Jul 19, 2023Abstract:In an effort to catalog insect biodiversity, we propose a new large dataset of hand-labelled insect images, the BIOSCAN-Insect Dataset. Each record is taxonomically classified by an expert, and also has associated genetic information including raw nucleotide barcode sequences and assigned barcode index numbers, which are genetically-based proxies for species classification. This paper presents a curated million-image dataset, primarily to train computer-vision models capable of providing image-based taxonomic assessment, however, the dataset also presents compelling characteristics, the study of which would be of interest to the broader machine learning community. Driven by the biological nature inherent to the dataset, a characteristic long-tailed class-imbalance distribution is exhibited. Furthermore, taxonomic labelling is a hierarchical classification scheme, presenting a highly fine-grained classification problem at lower levels. Beyond spurring interest in biodiversity research within the machine learning community, progress on creating an image-based taxonomic classifier will also further the ultimate goal of all BIOSCAN research: to lay the foundation for a comprehensive survey of global biodiversity. This paper introduces the dataset and explores the classification task through the implementation and analysis of a baseline classifier.
MOTCOM: The Multi-Object Tracking Dataset Complexity Metric
Jul 20, 2022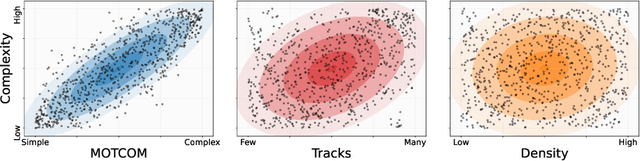
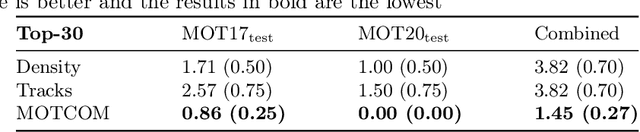
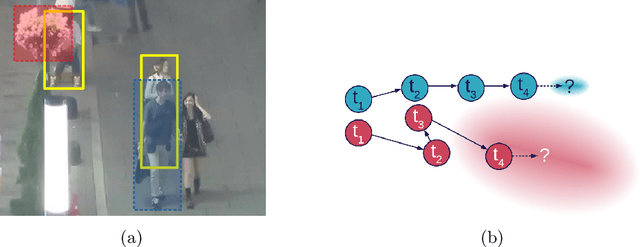

Abstract:There exists no comprehensive metric for describing the complexity of Multi-Object Tracking (MOT) sequences. This lack of metrics decreases explainability, complicates comparison of datasets, and reduces the conversation on tracker performance to a matter of leader board position. As a remedy, we present the novel MOT dataset complexity metric (MOTCOM), which is a combination of three sub-metrics inspired by key problems in MOT: occlusion, erratic motion, and visual similarity. The insights of MOTCOM can open nuanced discussions on tracker performance and may lead to a wider acknowledgement of novel contributions developed for either less known datasets or those aimed at solving sub-problems. We evaluate MOTCOM on the comprehensive MOT17, MOT20, and MOTSynth datasets and show that MOTCOM is far better at describing the complexity of MOT sequences compared to the conventional density and number of tracks. Project page at https://vap.aau.dk/motcom
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge