James E. D. Tweel
Photon Absorption Remote Sensing Virtual Histopathology: Diagnostic Equivalence to Gold-Standard H&E Staining in Skin Cancer Excisional Biopsies
Apr 25, 2025Abstract:Photon Absorption Remote Sensing (PARS) enables label-free imaging of subcellular morphology by observing biomolecule specific absorption interactions. Coupled with deep-learning, PARS produces label-free virtual Hematoxylin and Eosin (H&E) stained images in unprocessed tissues. This study evaluates the diagnostic performance of these PARS-derived virtual H&E images in benign and malignant excisional skin biopsies, including Squamous (SCC), Basal (BCC) Cell Carcinoma, and normal skin. Sixteen unstained formalin-fixed paraffin-embedded skin excisions were PARS imaged, virtually H&E stained, then chemically stained and imaged at 40x. Seven fellowship trained dermatopathologists assessed all 32 images in a masked randomized fashion. Concordance analysis indicates 95.5% agreement between primary diagnoses rendered on PARS versus H&E images (Cohen's k=0.93). Inter-rater reliability was near-perfect for both image types (Fleiss' k=0.89 for PARS, k=0.80 for H&E). For subtype classification, agreement was near-perfect 91% (k=0.73) for SCC and was perfect for BCC. When assessing malignancy confinement (e.g., cancer margins), agreement was 92% between PARS and H&E (k=0.718). During assessment dermatopathologists could not reliably distinguish image origin (PARS vs. H&E), and diagnostic confidence was equivalent between the modalities. Inter-rater reliability for PARS virtual H&E was consistent with reported benchmarks for histologic evaluation. These results indicate that PARS virtual histology may be diagnostically equivalent to traditional H&E staining in dermatopathology diagnostics, while enabling assessment directly from unlabeled, or unprocessed slides. In turn, the label-free PARS virtual H&E imaging workflow may preserve tissue for downstream analysis while producing data well-suited for AI integration potentially accelerating and enhancing the accuracy of skin cancer diagnostics.
Multi-Channel Feature Extraction for Virtual Histological Staining of Photon Absorption Remote Sensing Images
Jul 04, 2023Abstract:Accurate and fast histological staining is crucial in histopathology, impacting diagnostic precision and reliability. Traditional staining methods are time-consuming and subjective, causing delays in diagnosis. Digital pathology plays a vital role in advancing and optimizing histology processes to improve efficiency and reduce turnaround times. This study introduces a novel deep learning-based framework for virtual histological staining using photon absorption remote sensing (PARS) images. By extracting features from PARS time-resolved signals using a variant of the K-means method, valuable multi-modal information is captured. The proposed multi-channel cycleGAN (MC-GAN) model expands on the traditional cycleGAN framework, allowing the inclusion of additional features. Experimental results reveal that specific combinations of features outperform the conventional channels by improving the labeling of tissue structures prior to model training. Applied to human skin and mouse brain tissue, the results underscore the significance of choosing the optimal combination of features, as it reveals a substantial visual and quantitative concurrence between the virtually stained and the gold standard chemically stained hematoxylin and eosin (H&E) images, surpassing the performance of other feature combinations. Accurate virtual staining is valuable for reliable diagnostic information, aiding pathologists in disease classification, grading, and treatment planning. This study aims to advance label-free histological imaging and opens doors for intraoperative microscopy applications.
Virtual Histology with Photon Absorption Remote Sensing using a Cycle-Consistent Generative Adversarial Network with Weakly Registered Pairs
Jun 26, 2023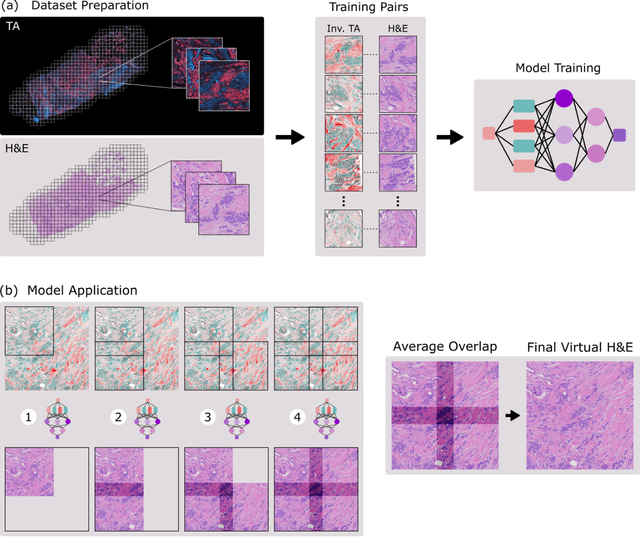
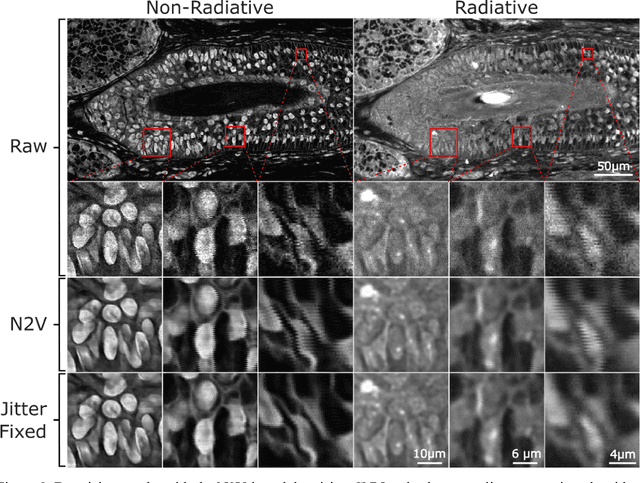
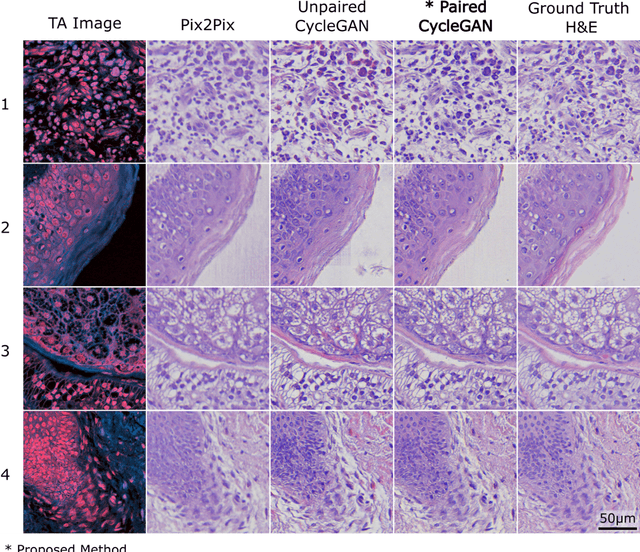
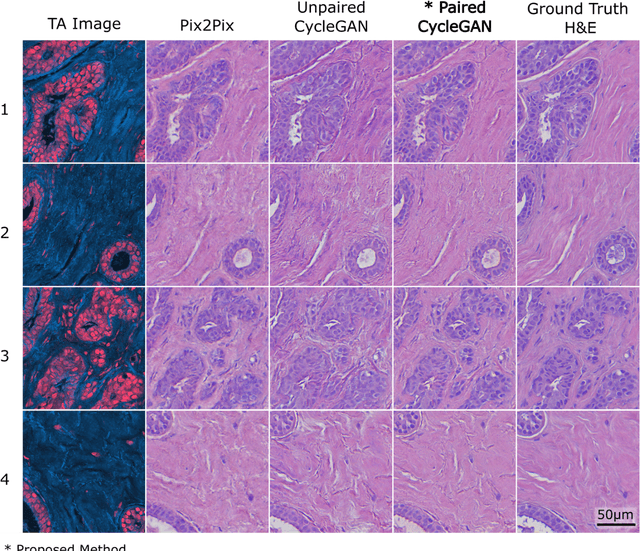
Abstract:Modern histopathology relies on the microscopic examination of thin tissue sections stained with histochemical techniques, typically using brightfield or fluorescence microscopy. However, the staining of samples can permanently alter their chemistry and structure, meaning an individual tissue section must be prepared for each desired staining contrast. This not only consumes valuable tissue samples but also introduces delays in essential diagnostic timelines. In this work, virtual histochemical staining is developed using label-free photon absorption remote sensing (PARS) microscopy. We present a method that generates virtually stained histology images that are indistinguishable from the gold standard hematoxylin and eosin (H&E) staining. First, PARS label-free ultraviolet absorption images are captured directly within unstained tissue specimens. The radiative and non-radiative absorption images are then preprocessed, and virtually stained through the presented pathway. The preprocessing pipeline features a self-supervised Noise2Void denoising convolutional neural network (CNN) as well as a novel algorithm for pixel-level mechanical scanning error correction. These developments significantly enhance the recovery of sub-micron tissue structures, such as nucleoli location and chromatin distribution. Finally, we used a cycle-consistent generative adversarial network CycleGAN architecture to virtually stain the preprocessed PARS data. Virtual staining is applied to thin unstained sections of malignant human skin and breast tissue samples. Clinically relevant details are revealed, with comparable contrast and quality to gold standard H&E-stained images. This work represents a crucial step to deploying label-free microscopy as an alternative to standard histopathology techniques.
Automated Whole Slide Imaging for Label-Free Histology using Photon Absorption Remote Sensing Microscopy
Apr 26, 2023Abstract:The field of histology relies heavily on antiquated tissue processing and staining techniques that limit the efficiency of pathologic diagnoses of cancer and other diseases. Current staining and advanced labeling methods are often destructive and mutually incompatible, requiring new tissue sections for each stain. This prolongs the diagnostic process and depletes valuable biopsy samples. In this study, we present an alternative label-free histology platform using the first transmission-mode Photon Absorption Remote Sensing microscope. Optimized for automated whole slide scanning of unstained tissue samples, the system provides slide images at magnifications up to 40x that are fully compatible with existing digital pathology tools. The scans capture high quality and high-resolution images with subcellular diagnostic detail. After imaging, samples remain suitable for histochemical, immunohistochemical, and other staining techniques. Scattering and absorption (radiative and non-radiative) contrasts are shown in whole slide images of malignant human breast and skin tissues samples. Clinically relevant features are highlighted, and close correspondence and analogous contrast is demonstrated with one-to-one gold standard H&E stained images. Our previously reported pix2pix virtual staining model is applied to an entire whole slide image, showcasing the potential of this approach in whole slide label-free H&E emulation. This work is a critical advance for integrating label-free optical methods into standard histopathology workflows, both enhancing diagnostic efficiency, and broadening the number of stains that can be applied while preserving valuable tissue samples.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge