Amir Borhani
Eyes Tell the Truth: GazeVal Highlights Shortcomings of Generative AI in Medical Imaging
Mar 26, 2025



Abstract:The demand for high-quality synthetic data for model training and augmentation has never been greater in medical imaging. However, current evaluations predominantly rely on computational metrics that fail to align with human expert recognition. This leads to synthetic images that may appear realistic numerically but lack clinical authenticity, posing significant challenges in ensuring the reliability and effectiveness of AI-driven medical tools. To address this gap, we introduce GazeVal, a practical framework that synergizes expert eye-tracking data with direct radiological evaluations to assess the quality of synthetic medical images. GazeVal leverages gaze patterns of radiologists as they provide a deeper understanding of how experts perceive and interact with synthetic data in different tasks (i.e., diagnostic or Turing tests). Experiments with sixteen radiologists revealed that 96.6% of the generated images (by the most recent state-of-the-art AI algorithm) were identified as fake, demonstrating the limitations of generative AI in producing clinically accurate images.
A Novel Momentum-Based Deep Learning Techniques for Medical Image Classification and Segmentation
Aug 11, 2024



Abstract:Accurately segmenting different organs from medical images is a critical prerequisite for computer-assisted diagnosis and intervention planning. This study proposes a deep learning-based approach for segmenting various organs from CT and MRI scans and classifying diseases. Our study introduces a novel technique integrating momentum within residual blocks for enhanced training dynamics in medical image analysis. We applied our method in two distinct tasks: segmenting liver, lung, & colon data and classifying abdominal pelvic CT and MRI scans. The proposed approach has shown promising results, outperforming state-of-the-art methods on publicly available benchmarking datasets. For instance, in the lung segmentation dataset, our approach yielded significant enhancements over the TransNetR model, including a 5.72% increase in dice score, a 5.04% improvement in mean Intersection over Union (mIoU), an 8.02% improvement in recall, and a 4.42% improvement in precision. Hence, incorporating momentum led to state-of-the-art performance in both segmentation and classification tasks, representing a significant advancement in the field of medical imaging.
MDNet: Multi-Decoder Network for Abdominal CT Organs Segmentation
May 10, 2024



Abstract:Accurate segmentation of organs from abdominal CT scans is essential for clinical applications such as diagnosis, treatment planning, and patient monitoring. To handle challenges of heterogeneity in organ shapes, sizes, and complex anatomical relationships, we propose a \textbf{\textit{\ac{MDNet}}}, an encoder-decoder network that uses the pre-trained \textit{MiT-B2} as the encoder and multiple different decoder networks. Each decoder network is connected to a different part of the encoder via a multi-scale feature enhancement dilated block. With each decoder, we increase the depth of the network iteratively and refine segmentation masks, enriching feature maps by integrating previous decoders' feature maps. To refine the feature map further, we also utilize the predicted masks from the previous decoder to the current decoder to provide spatial attention across foreground and background regions. MDNet effectively refines the segmentation mask with a high dice similarity coefficient (DSC) of 0.9013 and 0.9169 on the Liver Tumor segmentation (LiTS) and MSD Spleen datasets. Additionally, it reduces Hausdorff distance (HD) to 3.79 for the LiTS dataset and 2.26 for the spleen segmentation dataset, underscoring the precision of MDNet in capturing the complex contours. Moreover, \textit{\ac{MDNet}} is more interpretable and robust compared to the other baseline models.
PAM-UNet: Shifting Attention on Region of Interest in Medical Images
May 02, 2024



Abstract:Computer-aided segmentation methods can assist medical personnel in improving diagnostic outcomes. While recent advancements like UNet and its variants have shown promise, they face a critical challenge: balancing accuracy with computational efficiency. Shallow encoder architectures in UNets often struggle to capture crucial spatial features, leading in inaccurate and sparse segmentation. To address this limitation, we propose a novel \underline{P}rogressive \underline{A}ttention based \underline{M}obile \underline{UNet} (\underline{PAM-UNet}) architecture. The inverted residual (IR) blocks in PAM-UNet help maintain a lightweight framework, while layerwise \textit{Progressive Luong Attention} ($\mathcal{PLA}$) promotes precise segmentation by directing attention toward regions of interest during synthesis. Our approach prioritizes both accuracy and speed, achieving a commendable balance with a mean IoU of 74.65 and a dice score of 82.87, while requiring only 1.32 floating-point operations per second (FLOPS) on the Liver Tumor Segmentation Benchmark (LiTS) 2017 dataset. These results highlight the importance of developing efficient segmentation models to accelerate the adoption of AI in clinical practice.
Detection of Peri-Pancreatic Edema using Deep Learning and Radiomics Techniques
Apr 25, 2024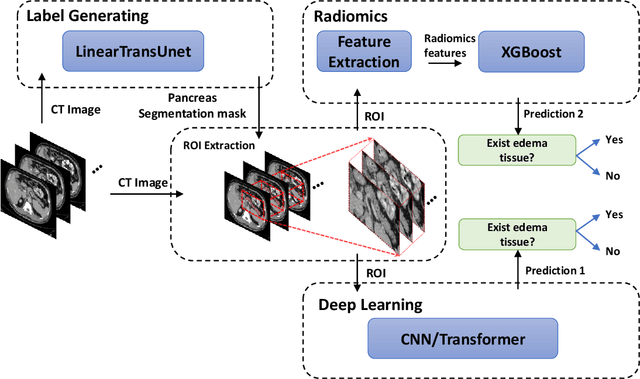
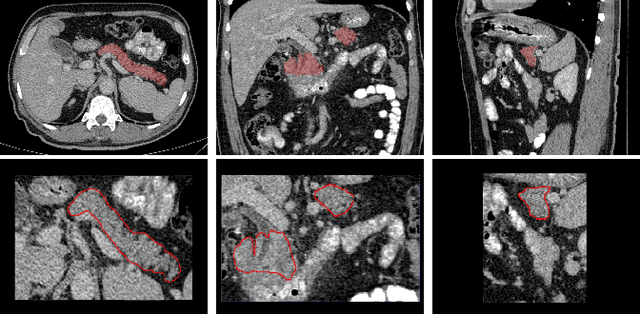
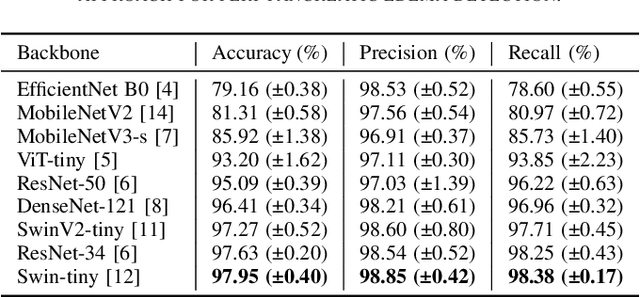
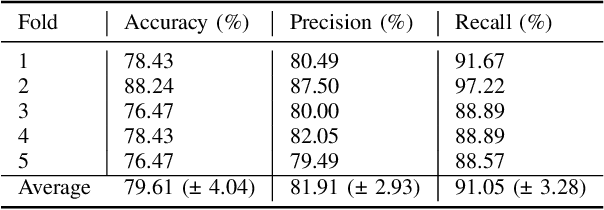
Abstract:Identifying peri-pancreatic edema is a pivotal indicator for identifying disease progression and prognosis, emphasizing the critical need for accurate detection and assessment in pancreatitis diagnosis and management. This study \textit{introduces a novel CT dataset sourced from 255 patients with pancreatic diseases, featuring annotated pancreas segmentation masks and corresponding diagnostic labels for peri-pancreatic edema condition}. With the novel dataset, we first evaluate the efficacy of the \textit{LinTransUNet} model, a linear Transformer based segmentation algorithm, to segment the pancreas accurately from CT imaging data. Then, we use segmented pancreas regions with two distinctive machine learning classifiers to identify existence of peri-pancreatic edema: deep learning-based models and a radiomics-based eXtreme Gradient Boosting (XGBoost). The LinTransUNet achieved promising results, with a dice coefficient of 80.85\%, and mIoU of 68.73\%. Among the nine benchmarked classification models for peri-pancreatic edema detection, \textit{Swin-Tiny} transformer model demonstrated the highest recall of $98.85 \pm 0.42$ and precision of $98.38\pm 0.17$. Comparatively, the radiomics-based XGBoost model achieved an accuracy of $79.61\pm4.04$ and recall of $91.05\pm3.28$, showcasing its potential as a supplementary diagnostic tool given its rapid processing speed and reduced training time. Our code is available \url{https://github.com/NUBagciLab/Peri-Pancreatic-Edema-Detection}.
CT Liver Segmentation via PVT-based Encoding and Refined Decoding
Jan 17, 2024


Abstract:Accurate liver segmentation from CT scans is essential for computer-aided diagnosis and treatment planning. Recently, Vision Transformers achieved a competitive performance in computer vision tasks compared to convolutional neural networks due to their exceptional ability to learn global representations. However, they often struggle with scalability, memory constraints, and computational inefficiency, particularly in handling high-resolution medical images. To overcome scalability and efficiency issues, we propose a novel deep learning approach, \textit{\textbf{PVTFormer}}, that is built upon a pretrained pyramid vision transformer (PVT v2) combined with advanced residual upsampling and decoder block. By integrating a refined feature channel approach with hierarchical decoding strategy, PVTFormer generates high quality segmentation masks by enhancing semantic features. Rigorous evaluation of the proposed method on Liver Tumor Segmentation Benchmark (LiTS) 2017 demonstrates that our proposed architecture not only achieves a high dice coefficient of 86.78\%, mIoU of 78.46\%, but also obtains a low HD of 3.50. The results underscore PVTFormer's efficacy in setting a new benchmark for state-of-the-art liver segmentation methods. The source code of the proposed PVTFormer is available at \url{https://github.com/DebeshJha/PVTFormer}.
Transformer based Generative Adversarial Network for Liver Segmentation
May 28, 2022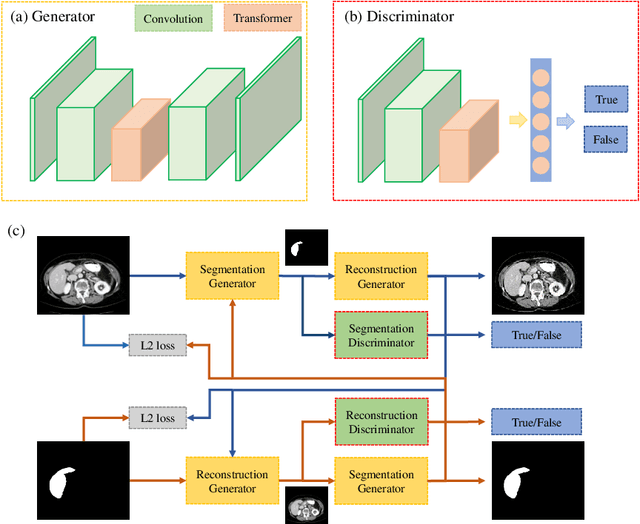
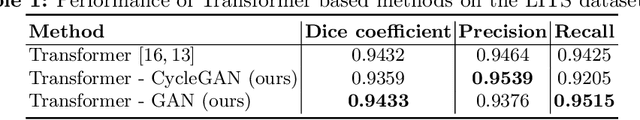
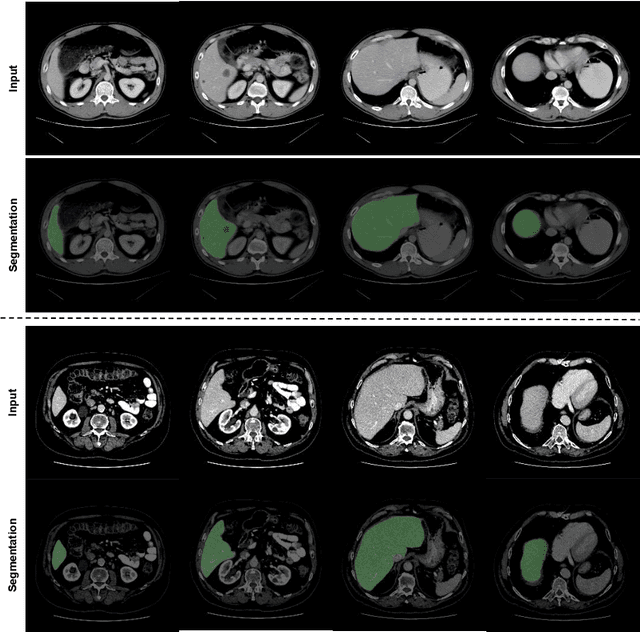
Abstract:Automated liver segmentation from radiology scans (CT, MRI) can improve surgery and therapy planning and follow-up assessment in addition to conventional use for diagnosis and prognosis. Although convolutional neural networks (CNNs) have become the standard image segmentation tasks, more recently this has started to change towards Transformers based architectures because Transformers are taking advantage of capturing long range dependence modeling capability in signals, so called attention mechanism. In this study, we propose a new segmentation approach using a hybrid approach combining the Transformer(s) with the Generative Adversarial Network (GAN) approach. The premise behind this choice is that the self-attention mechanism of the Transformers allows the network to aggregate the high dimensional feature and provide global information modeling. This mechanism provides better segmentation performance compared with traditional methods. Furthermore, we encode this generator into the GAN based architecture so that the discriminator network in the GAN can classify the credibility of the generated segmentation masks compared with the real masks coming from human (expert) annotations. This allows us to extract the high dimensional topology information in the mask for biomedical image segmentation and provide more reliable segmentation results. Our model achieved a high dice coefficient of 0.9433, recall of 0.9515, and precision of 0.9376 and outperformed other Transformer based approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge