Md Mostafijur Rahman
PipeFlow: Pipelined Processing and Motion-Aware Frame Selection for Long-Form Video Editing
Dec 30, 2025Abstract:Long-form video editing poses unique challenges due to the exponential increase in the computational cost from joint editing and Denoising Diffusion Implicit Models (DDIM) inversion across extended sequences. To address these limitations, we propose PipeFlow, a scalable, pipelined video editing method that introduces three key innovations: First, based on a motion analysis using Structural Similarity Index Measure (SSIM) and Optical Flow, we identify and propose to skip editing of frames with low motion. Second, we propose a pipelined task scheduling algorithm that splits a video into multiple segments and performs DDIM inversion and joint editing in parallel based on available GPU memory. Lastly, we leverage a neural network-based interpolation technique to smooth out the border frames between segments and interpolate the previously skipped frames. Our method uniquely scales to longer videos by dividing them into smaller segments, allowing PipeFlow's editing time to increase linearly with video length. In principle, this enables editing of infinitely long videos without the growing per-frame computational overhead encountered by other methods. PipeFlow achieves up to a 9.6X speedup compared to TokenFlow and a 31.7X speedup over Diffusion Motion Transfer (DMT).
AdaptViG: Adaptive Vision GNN with Exponential Decay Gating
Nov 13, 2025



Abstract:Vision Graph Neural Networks (ViGs) offer a new direction for advancements in vision architectures. While powerful, ViGs often face substantial computational challenges stemming from their graph construction phase, which can hinder their efficiency. To address this issue we propose AdaptViG, an efficient and powerful hybrid Vision GNN that introduces a novel graph construction mechanism called Adaptive Graph Convolution. This mechanism builds upon a highly efficient static axial scaffold and a dynamic, content-aware gating strategy called Exponential Decay Gating. This gating mechanism selectively weighs long-range connections based on feature similarity. Furthermore, AdaptViG employs a hybrid strategy, utilizing our efficient gating mechanism in the early stages and a full Global Attention block in the final stage for maximum feature aggregation. Our method achieves a new state-of-the-art trade-off between accuracy and efficiency among Vision GNNs. For instance, our AdaptViG-M achieves 82.6% top-1 accuracy, outperforming ViG-B by 0.3% while using 80% fewer parameters and 84% fewer GMACs. On downstream tasks, AdaptViG-M obtains 45.8 mIoU, 44.8 APbox, and 41.1 APmask, surpassing the much larger EfficientFormer-L7 by 0.7 mIoU, 2.2 APbox, and 2.1 APmask, respectively, with 78% fewer parameters.
LoMix: Learnable Weighted Multi-Scale Logits Mixing for Medical Image Segmentation
Oct 27, 2025Abstract:U-shaped networks output logits at multiple spatial scales, each capturing a different blend of coarse context and fine detail. Yet, training still treats these logits in isolation - either supervising only the final, highest-resolution logits or applying deep supervision with identical loss weights at every scale - without exploring mixed-scale combinations. Consequently, the decoder output misses the complementary cues that arise only when coarse and fine predictions are fused. To address this issue, we introduce LoMix (Logits Mixing), a NAS-inspired, differentiable plug-and-play module that generates new mixed-scale outputs and learns how exactly each of them should guide the training process. More precisely, LoMix mixes the multi-scale decoder logits with four lightweight fusion operators: addition, multiplication, concatenation, and attention-based weighted fusion, yielding a rich set of synthetic mutant maps. Every original or mutant map is given a softplus loss weight that is co-optimized with network parameters, mimicking a one-step architecture search that automatically discovers the most useful scales, mixtures, and operators. Plugging LoMix into recent U-shaped architectures (i.e., PVT-V2-B2 backbone with EMCAD decoder) on Synapse 8-organ dataset improves DICE by +4.2% over single-output supervision, +2.2% over deep supervision, and +1.5% over equally weighted additive fusion, all with zero inference overhead. When training data are scarce (e.g., one or two labeled scans), the advantage grows to +9.23%, underscoring LoMix's data efficiency. Across four benchmarks and diverse U-shaped networks, LoMiX improves DICE by up to +13.5% over single-output supervision, confirming that learnable weighted mixed-scale fusion generalizes broadly while remaining data efficient, fully interpretable, and overhead-free at inference. Our code is available at https://github.com/SLDGroup/LoMix.
An Efficient Dual-Line Decoder Network with Multi-Scale Convolutional Attention for Multi-organ Segmentation
Aug 23, 2025Abstract:Proper segmentation of organs-at-risk is important for radiation therapy, surgical planning, and diagnostic decision-making in medical image analysis. While deep learning-based segmentation architectures have made significant progress, they often fail to balance segmentation accuracy with computational efficiency. Most of the current state-of-the-art methods either prioritize performance at the cost of high computational complexity or compromise accuracy for efficiency. This paper addresses this gap by introducing an efficient dual-line decoder segmentation network (EDLDNet). The proposed method features a noisy decoder, which learns to incorporate structured perturbation at training time for better model robustness, yet at inference time only the noise-free decoder is executed, leading to lower computational cost. Multi-Scale convolutional Attention Modules (MSCAMs), Attention Gates (AGs), and Up-Convolution Blocks (UCBs) are further utilized to optimize feature representation and boost segmentation performance. By leveraging multi-scale segmentation masks from both decoders, we also utilize a mutation-based loss function to enhance the model's generalization. Our approach outperforms SOTA segmentation architectures on four publicly available medical imaging datasets. EDLDNet achieves SOTA performance with an 84.00% Dice score on the Synapse dataset, surpassing baseline model like UNet by 13.89% in Dice score while significantly reducing Multiply-Accumulate Operations (MACs) by 89.7%. Compared to recent approaches like EMCAD, our EDLDNet not only achieves higher Dice score but also maintains comparable computational efficiency. The outstanding performance across diverse datasets establishes EDLDNet's strong generalization, computational efficiency, and robustness. The source code, pre-processed data, and pre-trained weights will be available at https://github.com/riadhassan/EDLDNet .
RapidNet: Multi-Level Dilated Convolution Based Mobile Backbone
Dec 14, 2024Abstract:Vision transformers (ViTs) have dominated computer vision in recent years. However, ViTs are computationally expensive and not well suited for mobile devices; this led to the prevalence of convolutional neural network (CNN) and ViT-based hybrid models for mobile vision applications. Recently, Vision GNN (ViG) and CNN hybrid models have also been proposed for mobile vision tasks. However, all of these methods remain slower compared to pure CNN-based models. In this work, we propose Multi-Level Dilated Convolutions to devise a purely CNN-based mobile backbone. Using Multi-Level Dilated Convolutions allows for a larger theoretical receptive field than standard convolutions. Different levels of dilation also allow for interactions between the short-range and long-range features in an image. Experiments show that our proposed model outperforms state-of-the-art (SOTA) mobile CNN, ViT, ViG, and hybrid architectures in terms of accuracy and/or speed on image classification, object detection, instance segmentation, and semantic segmentation. Our fastest model, RapidNet-Ti, achieves 76.3\% top-1 accuracy on ImageNet-1K with 0.9 ms inference latency on an iPhone 13 mini NPU, which is faster and more accurate than MobileNetV2x1.4 (74.7\% top-1 with 1.0 ms latency). Our work shows that pure CNN architectures can beat SOTA hybrid and ViT models in terms of accuracy and speed when designed properly.
PP-SAM: Perturbed Prompts for Robust Adaptation of Segment Anything Model for Polyp Segmentation
May 27, 2024Abstract:The Segment Anything Model (SAM), originally designed for general-purpose segmentation tasks, has been used recently for polyp segmentation. Nonetheless, fine-tuning SAM with data from new imaging centers or clinics poses significant challenges. This is because this necessitates the creation of an expensive and time-intensive annotated dataset, along with the potential for variability in user prompts during inference. To address these issues, we propose a robust fine-tuning technique, PP-SAM, that allows SAM to adapt to the polyp segmentation task with limited images. To this end, we utilize variable perturbed bounding box prompts (BBP) to enrich the learning context and enhance the model's robustness to BBP perturbations during inference. Rigorous experiments on polyp segmentation benchmarks reveal that our variable BBP perturbation significantly improves model resilience. Notably, on Kvasir, 1-shot fine-tuning boosts the DICE score by 20% and 37% with 50 and 100-pixel BBP perturbations during inference, respectively. Moreover, our experiments show that 1-shot, 5-shot, and 10-shot PP-SAM with 50-pixel perturbations during inference outperform a recent state-of-the-art (SOTA) polyp segmentation method by 26%, 7%, and 5% DICE scores, respectively. Our results motivate the broader applicability of our PP-SAM for other medical imaging tasks with limited samples. Our implementation is available at https://github.com/SLDGroup/PP-SAM.
EMCAD: Efficient Multi-scale Convolutional Attention Decoding for Medical Image Segmentation
May 11, 2024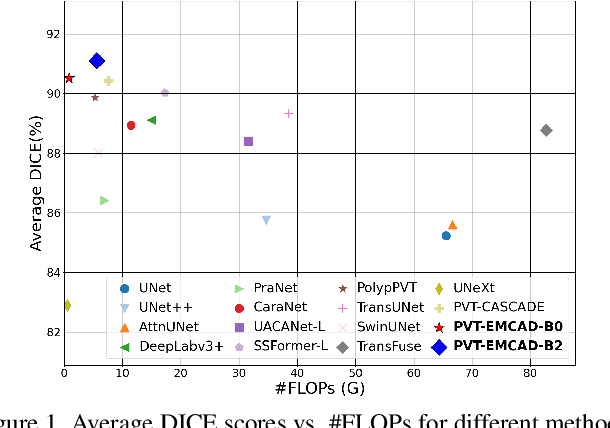
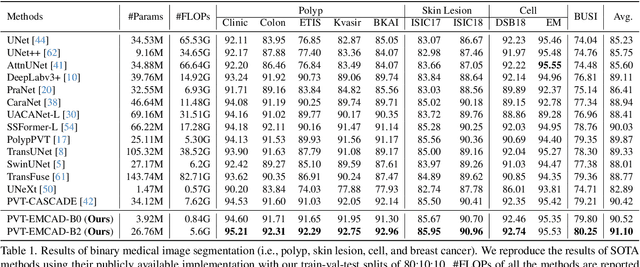
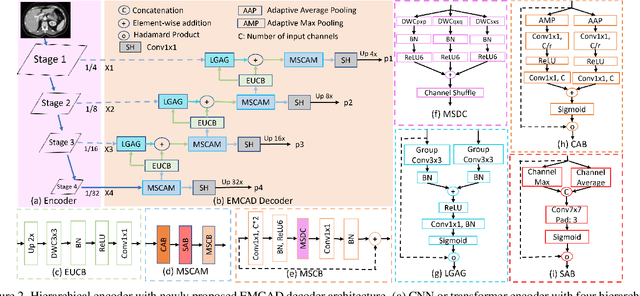
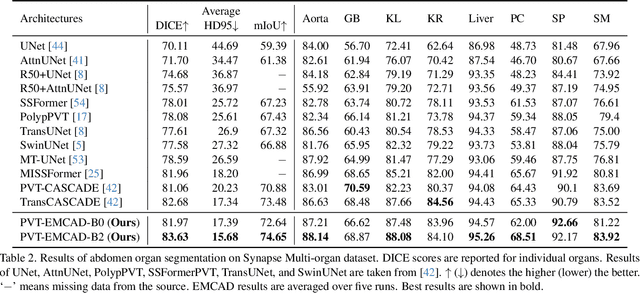
Abstract:An efficient and effective decoding mechanism is crucial in medical image segmentation, especially in scenarios with limited computational resources. However, these decoding mechanisms usually come with high computational costs. To address this concern, we introduce EMCAD, a new efficient multi-scale convolutional attention decoder, designed to optimize both performance and computational efficiency. EMCAD leverages a unique multi-scale depth-wise convolution block, significantly enhancing feature maps through multi-scale convolutions. EMCAD also employs channel, spatial, and grouped (large-kernel) gated attention mechanisms, which are highly effective at capturing intricate spatial relationships while focusing on salient regions. By employing group and depth-wise convolution, EMCAD is very efficient and scales well (e.g., only 1.91M parameters and 0.381G FLOPs are needed when using a standard encoder). Our rigorous evaluations across 12 datasets that belong to six medical image segmentation tasks reveal that EMCAD achieves state-of-the-art (SOTA) performance with 79.4% and 80.3% reduction in #Params and #FLOPs, respectively. Moreover, EMCAD's adaptability to different encoders and versatility across segmentation tasks further establish EMCAD as a promising tool, advancing the field towards more efficient and accurate medical image analysis. Our implementation is available at https://github.com/SLDGroup/EMCAD.
MDNet: Multi-Decoder Network for Abdominal CT Organs Segmentation
May 10, 2024



Abstract:Accurate segmentation of organs from abdominal CT scans is essential for clinical applications such as diagnosis, treatment planning, and patient monitoring. To handle challenges of heterogeneity in organ shapes, sizes, and complex anatomical relationships, we propose a \textbf{\textit{\ac{MDNet}}}, an encoder-decoder network that uses the pre-trained \textit{MiT-B2} as the encoder and multiple different decoder networks. Each decoder network is connected to a different part of the encoder via a multi-scale feature enhancement dilated block. With each decoder, we increase the depth of the network iteratively and refine segmentation masks, enriching feature maps by integrating previous decoders' feature maps. To refine the feature map further, we also utilize the predicted masks from the previous decoder to the current decoder to provide spatial attention across foreground and background regions. MDNet effectively refines the segmentation mask with a high dice similarity coefficient (DSC) of 0.9013 and 0.9169 on the Liver Tumor segmentation (LiTS) and MSD Spleen datasets. Additionally, it reduces Hausdorff distance (HD) to 3.79 for the LiTS dataset and 2.26 for the spleen segmentation dataset, underscoring the precision of MDNet in capturing the complex contours. Moreover, \textit{\ac{MDNet}} is more interpretable and robust compared to the other baseline models.
GreedyViG: Dynamic Axial Graph Construction for Efficient Vision GNNs
May 10, 2024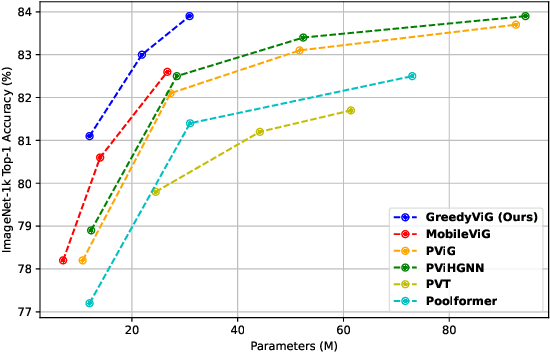
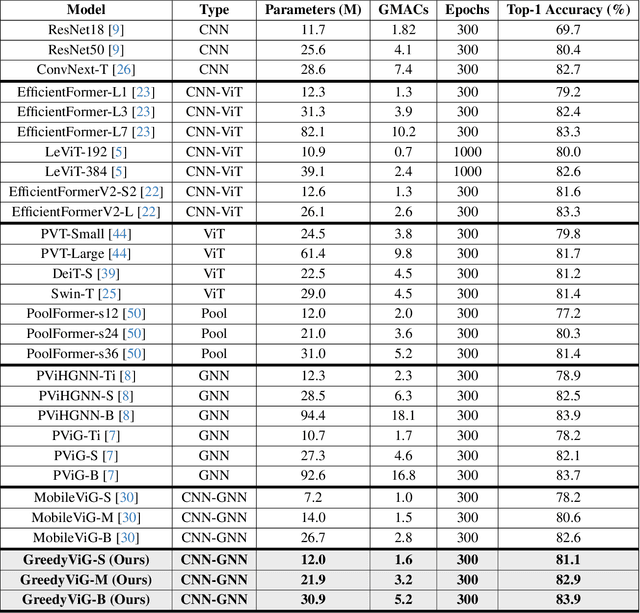
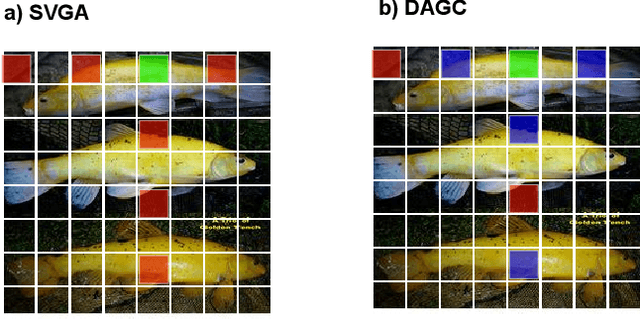
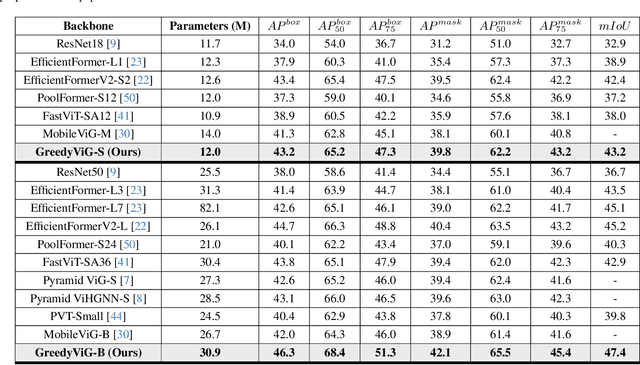
Abstract:Vision graph neural networks (ViG) offer a new avenue for exploration in computer vision. A major bottleneck in ViGs is the inefficient k-nearest neighbor (KNN) operation used for graph construction. To solve this issue, we propose a new method for designing ViGs, Dynamic Axial Graph Construction (DAGC), which is more efficient than KNN as it limits the number of considered graph connections made within an image. Additionally, we propose a novel CNN-GNN architecture, GreedyViG, which uses DAGC. Extensive experiments show that GreedyViG beats existing ViG, CNN, and ViT architectures in terms of accuracy, GMACs, and parameters on image classification, object detection, instance segmentation, and semantic segmentation tasks. Our smallest model, GreedyViG-S, achieves 81.1% top-1 accuracy on ImageNet-1K, 2.9% higher than Vision GNN and 2.2% higher than Vision HyperGraph Neural Network (ViHGNN), with less GMACs and a similar number of parameters. Our largest model, GreedyViG-B obtains 83.9% top-1 accuracy, 0.2% higher than Vision GNN, with a 66.6% decrease in parameters and a 69% decrease in GMACs. GreedyViG-B also obtains the same accuracy as ViHGNN with a 67.3% decrease in parameters and a 71.3% decrease in GMACs. Our work shows that hybrid CNN-GNN architectures not only provide a new avenue for designing efficient models, but that they can also exceed the performance of current state-of-the-art models.
BanglaNet: Bangla Handwritten Character Recognition using Ensembling of Convolutional Neural Network
Feb 04, 2024Abstract:Handwritten character recognition is a crucial task because of its abundant applications. The recognition task of Bangla handwritten characters is especially challenging because of the cursive nature of Bangla characters and the presence of compound characters with more than one way of writing. In this paper, a classification model based on the ensembling of several Convolutional Neural Networks (CNN), namely, BanglaNet is proposed to classify Bangla basic characters, compound characters, numerals, and modifiers. Three different models based on the idea of state-of-the-art CNN models like Inception, ResNet, and DenseNet have been trained with both augmented and non-augmented inputs. Finally, all these models are averaged or ensembled to get the finishing model. Rigorous experimentation on three benchmark Bangla handwritten characters datasets, namely, CMATERdb, BanglaLekha-Isolated, and Ekush has exhibited significant recognition accuracies compared to some recent CNN-based research. The top-1 recognition accuracies obtained are 98.40%, 97.65%, and 97.32%, and the top-3 accuracies are 99.79%, 99.74%, and 99.56% for CMATERdb, BanglaLekha-Isolated, and Ekush datasets respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge