Ismail Irmakci
Multi-Contrast MRI Segmentation Trained on Synthetic Images
Jul 06, 2022
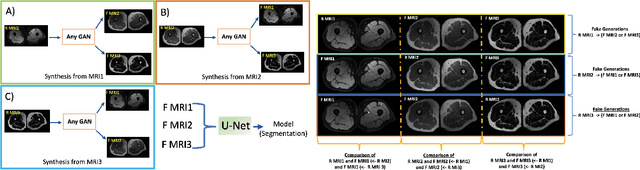

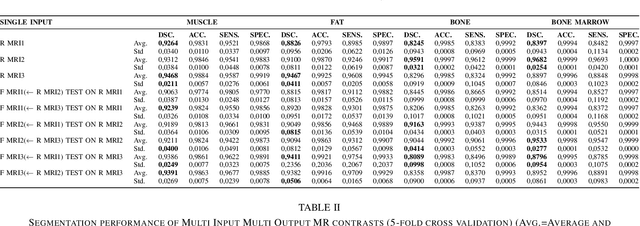
Abstract:In our comprehensive experiments and evaluations, we show that it is possible to generate multiple contrast (even all synthetically) and use synthetically generated images to train an image segmentation engine. We showed promising segmentation results tested on real multi-contrast MRI scans when delineating muscle, fat, bone and bone marrow, all trained on synthetic images. Based on synthetic image training, our segmentation results were as high as 93.91\%, 94.11\%, 91.63\%, 95.33\%, for muscle, fat, bone, and bone marrow delineation, respectively. Results were not significantly different from the ones obtained when real images were used for segmentation training: 94.68\%, 94.67\%, 95.91\%, and 96.82\%, respectively.
Neural Transformers for Intraductal Papillary Mucosal Neoplasms (IPMN) Classification in MRI images
Jun 21, 2022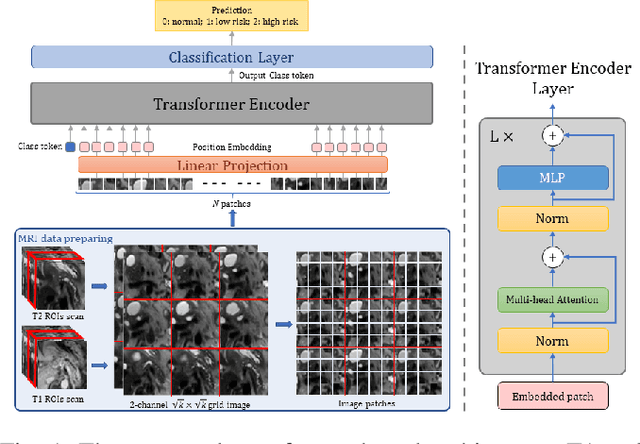
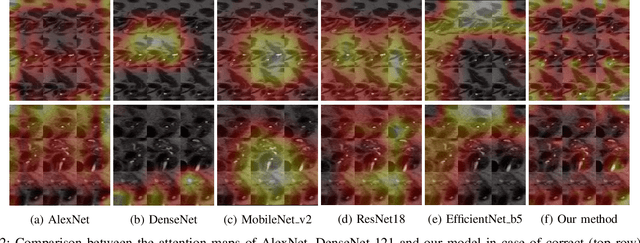

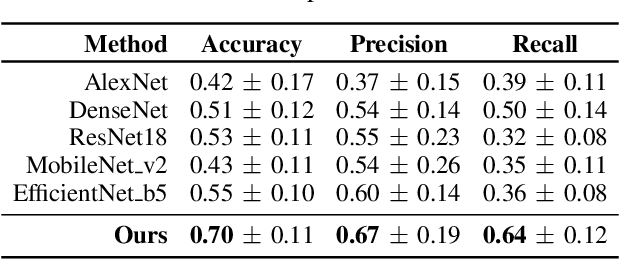
Abstract:Early detection of precancerous cysts or neoplasms, i.e., Intraductal Papillary Mucosal Neoplasms (IPMN), in pancreas is a challenging and complex task, and it may lead to a more favourable outcome. Once detected, grading IPMNs accurately is also necessary, since low-risk IPMNs can be under surveillance program, while high-risk IPMNs have to be surgically resected before they turn into cancer. Current standards (Fukuoka and others) for IPMN classification show significant intra- and inter-operator variability, beside being error-prone, making a proper diagnosis unreliable. The established progress in artificial intelligence, through the deep learning paradigm, may provide a key tool for an effective support to medical decision for pancreatic cancer. In this work, we follow this trend, by proposing a novel AI-based IPMN classifier that leverages the recent success of transformer networks in generalizing across a wide variety of tasks, including vision ones. We specifically show that our transformer-based model exploits pre-training better than standard convolutional neural networks, thus supporting the sought architectural universalism of transformers in vision, including the medical image domain and it allows for a better interpretation of the obtained results.
Hierarchical 3D Feature Learning for Pancreas Segmentation
Sep 03, 2021
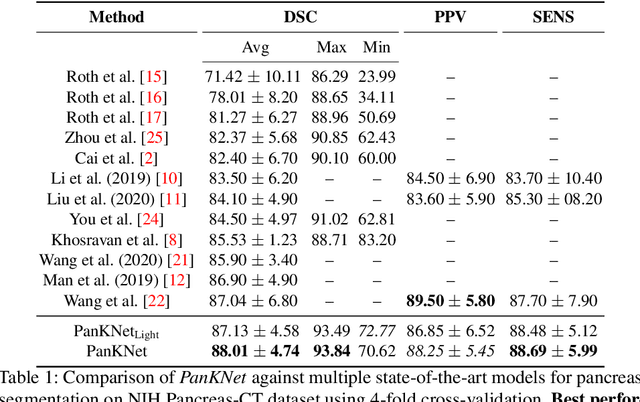
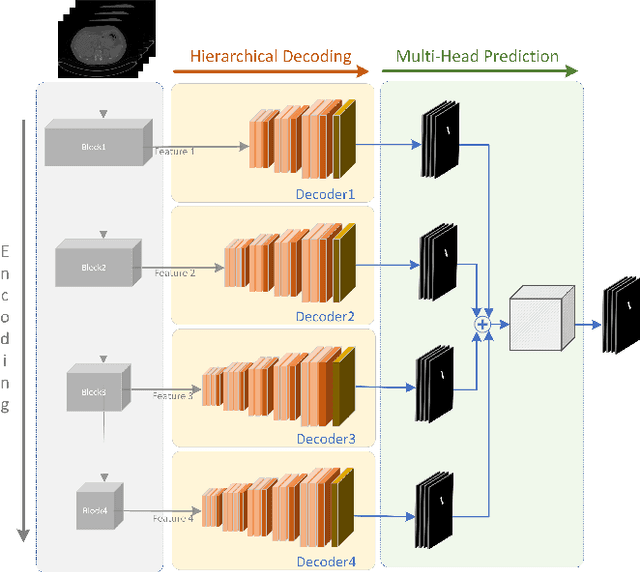

Abstract:We propose a novel 3D fully convolutional deep network for automated pancreas segmentation from both MRI and CT scans. More specifically, the proposed model consists of a 3D encoder that learns to extract volume features at different scales; features taken at different points of the encoder hierarchy are then sent to multiple 3D decoders that individually predict intermediate segmentation maps. Finally, all segmentation maps are combined to obtain a unique detailed segmentation mask. We test our model on both CT and MRI imaging data: the publicly available NIH Pancreas-CT dataset (consisting of 82 contrast-enhanced CTs) and a private MRI dataset (consisting of 40 MRI scans). Experimental results show that our model outperforms existing methods on CT pancreas segmentation, obtaining an average Dice score of about 88%, and yields promising segmentation performance on a very challenging MRI data set (average Dice score is about 77%). Additional control experiments demonstrate that the achieved performance is due to the combination of our 3D fully-convolutional deep network and the hierarchical representation decoding, thus substantiating our architectural design.
Information Bottleneck Attribution for Visual Explanations of Diagnosis and Prognosis
Apr 07, 2021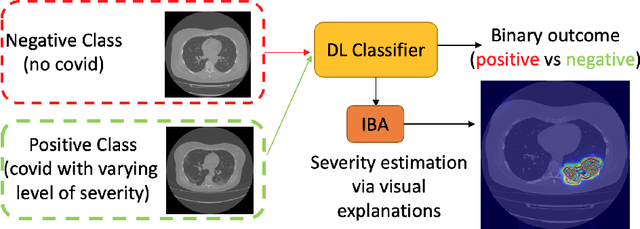



Abstract:Visual explanation methods have an important role in the prognosis of the patients where the annotated data is limited or not available. There have been several attempts to use gradient-based attribution methods to localize pathology from medical scans without using segmentation labels. This research direction has been impeded by the lack of robustness and reliability. These methods are highly sensitive to the network parameters. In this study, we introduce a robust visual explanation method to address this problem for medical applications. We provide a highly innovative algorithm to quantifying lesions in the lungs caused by the Covid-19 with high accuracy and robustness without using dense segmentation labels. Inspired by the information bottleneck concept, we mask the neural network representation with noise to find out important regions. This approach overcomes the drawbacks of commonly used Grad-Cam and its derived algorithms. The premise behind our proposed strategy is that the information flow is minimized while ensuring the classifier prediction stays similar. Our findings indicate that the bottleneck condition provides a more stable and robust severity estimation than the similar attribution methods.
Deep Learning for Musculoskeletal Image Analysis
Mar 01, 2020

Abstract:The diagnosis, prognosis, and treatment of patients with musculoskeletal (MSK) disorders require radiology imaging (using computed tomography, magnetic resonance imaging(MRI), and ultrasound) and their precise analysis by expert radiologists. Radiology scans can also help assessment of metabolic health, aging, and diabetes. This study presents how machinelearning, specifically deep learning methods, can be used for rapidand accurate image analysis of MRI scans, an unmet clinicalneed in MSK radiology. As a challenging example, we focus on automatic analysis of knee images from MRI scans and study machine learning classification of various abnormalities including meniscus and anterior cruciate ligament tears. Using widely used convolutional neural network (CNN) based architectures, we comparatively evaluated the knee abnormality classification performances of different neural network architectures under limited imaging data regime and compared single and multi-view imaging when classifying the abnormalities. Promising results indicated the potential use of multi-view deep learning based classification of MSK abnormalities in routine clinical assessment.
A Novel Extension to Fuzzy Connectivity for Body Composition Analysis: Applications in Thigh, Brain, and Whole Body Tissue Segmentation
Oct 14, 2018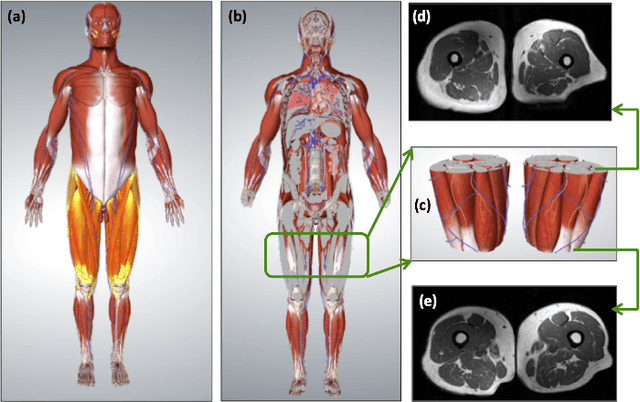
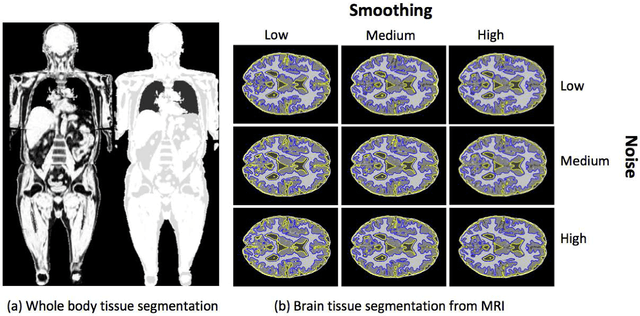
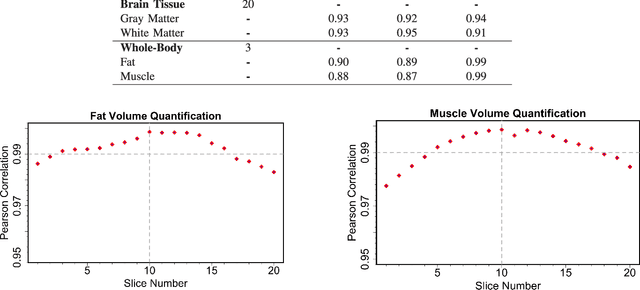
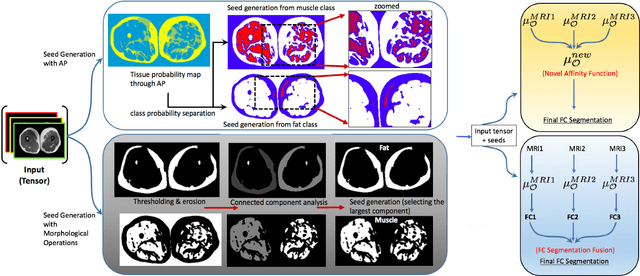
Abstract:Magnetic resonance imaging (MRI) is the non-invasive modality of choice for body tissue composition analysis due to its excellent soft tissue contrast and lack of ionizing radiation. However, quantification of body composition requires an accurate segmentation of fat, muscle and other tissues from MR images, which remains a challenging goal due to the intensity overlap between them. In this study, we propose a fully automated, data-driven image segmentation platform that addresses multiple difficulties in segmenting MR images such as varying inhomogeneity, non-standardness, and noise, while producing high-quality definition of different tissues. In contrast to most approaches in the literature, we perform segmentation operation by combining three different MRI contrasts and a novel segmentation tool which takes into account variability in the data. The proposed system, based on a novel affinity definition within the fuzzy connectivity (FC) image segmentation family, prevents the need for user intervention and reparametrization of the segmentation algorithms. In order to make the whole system fully automated, we adapt an affinity propagation clustering algorithm to roughly identify tissue regions and image background. We perform a thorough evaluation of the proposed algorithm's individual steps as well as comparison with several approaches from the literature for the main application of muscle/fat separation. Furthermore, whole-body tissue composition and brain tissue delineation were conducted to show the generalization ability of the proposed system. This new automated platform outperforms other state-of-the-art segmentation approaches both in accuracy and efficiency.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge